+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the SbCas7-11-crRNA-NTR complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / RNA-binding / RNA BINDING PROTEIN | |||||||||
| Function / homology | : / CRISPR type III-associated protein / RAMP superfamily / defense response to virus / RAMP superfamily protein Function and homology information Function and homology information | |||||||||
| Biological species |  Candidatus Scalindua brodae (bacteria) Candidatus Scalindua brodae (bacteria) | |||||||||
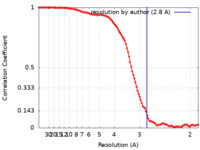
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Yu G / Wang X / Deng Z / Zhang H | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2022 Journal: Nucleic Acids Res / Year: 2022Title: Target RNA-guided protease activity in type III-E CRISPR-Cas system. Authors: Xiaoshen Wang / Guimei Yu / Yanan Wen / Qiyin An / Xuzichao Li / Fumeng Liao / Chengwei Lian / Kai Zhang / Hang Yin / Yong Wei / Zengqin Deng / Heng Zhang /  Abstract: The type III-E CRISPR-Cas systems are newly identified adaptive immune systems in prokaryotes that use a single Cas7-11 protein to specifically cleave target RNA. Cas7-11 could associate with Csx29, ...The type III-E CRISPR-Cas systems are newly identified adaptive immune systems in prokaryotes that use a single Cas7-11 protein to specifically cleave target RNA. Cas7-11 could associate with Csx29, a putative caspase-like protein encoded by the gene frequently found in the type III-E loci, suggesting a functional linkage between the RNase and protease activities in type III-E systems. Here, we demonstrated that target RNA recognition would stimulate the proteolytic activity of Csx29, and protein Csx30 is the endogenous substrate. More interestingly, while the cognate target RNA recognition would activate Csx29, non-cognate target RNA with the complementary 3' anti-tag sequence inhibits the enzymatic activity. Csx30 could bind to the sigma factor RpoE, which may initiate the stress response after proteolytic cleavage. Combined with biochemical and structural studies, we have elucidated the mechanisms underlying the target RNA-guided proteolytic activity of Csx29. Our work will guide further developments leveraging this simple RNA targeting system for RNA and protein-related applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34158.map.gz emd_34158.map.gz | 89.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34158-v30.xml emd-34158-v30.xml emd-34158.xml emd-34158.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34158_fsc.xml emd_34158_fsc.xml | 13.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_34158.png emd_34158.png | 118.9 KB | ||
| Filedesc metadata |  emd-34158.cif.gz emd-34158.cif.gz | 7.2 KB | ||
| Others |  emd_34158_half_map_1.map.gz emd_34158_half_map_1.map.gz emd_34158_half_map_2.map.gz emd_34158_half_map_2.map.gz | 8 MB 8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34158 http://ftp.pdbj.org/pub/emdb/structures/EMD-34158 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34158 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34158 | HTTPS FTP |
-Related structure data
| Related structure data |  8gnaMC  8gu6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34158.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34158.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.95 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34158_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_34158_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ternary complex
| Entire | Name: ternary complex |
|---|---|
| Components |
|
-Supramolecule #1: ternary complex
| Supramolecule | Name: ternary complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Candidatus Scalindua brodae (bacteria) Candidatus Scalindua brodae (bacteria) |
-Macromolecule #1: RAMP superfamily protein
| Macromolecule | Name: RAMP superfamily protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Candidatus Scalindua brodae (bacteria) Candidatus Scalindua brodae (bacteria) |
| Molecular weight | Theoretical: 197.173125 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNITVELTFF EPYRLVEWFD WDARKKSHSA MRGQAFAQWT WKGKGRTAGK SFITGTLVRS AVIKAVEELL SLNNGKWEGV PCCNGSFQT DESKGKKPSF LRKRHTLQWQ ANNKNICDKE EACPFCILLG RFDNAGKVHE RNKDYDIHFS NFDLDHKQEK N DLRLVDIA ...String: MNITVELTFF EPYRLVEWFD WDARKKSHSA MRGQAFAQWT WKGKGRTAGK SFITGTLVRS AVIKAVEELL SLNNGKWEGV PCCNGSFQT DESKGKKPSF LRKRHTLQWQ ANNKNICDKE EACPFCILLG RFDNAGKVHE RNKDYDIHFS NFDLDHKQEK N DLRLVDIA SGRILNRVDF DTGKAKDYFR TWEADYETYG TYTGRITLRN EHAKKLLLAS LGFVDKLCGA LCRIEVIKKS ES PLPSDTK EQSYTKDDTV EVLSEDHNDE LRKQAEVIVE AFKQNDKLEK IRILADAIRT LRLHGEGVIE KDELPDGKEE RDK GHHLWD IKVQGTALRT KLKELWQSNK DIGWRKFTEM LGSNLYLIYK KETGGVSTRF RILGDTEYYS KAHDSEGSDL FIPV TPPEG IETKEWIIVG RLKAATPFYF GVQQPSDSIP GKEKKSEDSL VINEHASFNI LLDKENRYRI PRSALRGALR RDLRT AFGS GCNVSLGGQI LCNCKVCIEM RRITLKDSVS DFSEPPEIRY RIAKNPGTAT VEDGSLFDIE VGPEGLTFPF VLRYRG HKF PEQLSSVIRY WEENDGKNGM AWLGGLDSTG KGRFALKDIK IFEWDLNQKI NEYIKERGMR GKEKELLEMG ESSLPDG LI PYKFFEEREC LFPYKENLKP QWSEVQYTIE VGSPLLTADT ISALTEPGNR AAIAYKKRVY NDGNNAIEPE PRFAVKSE T HRGIFRTAVG RRTGDLGKED HEDCTCDMCI IFGNEHESSK IRFEDLELIN GNEFEKLEKH IDHVAIDRFT GGALDKAKF DTYPLAGSPK KPLKLKGRFW IKKGFSGDHK LLITTALSDI RDGLYPLGSK GGVGYGWVAG ISIDDNVPDD FKEMINKTEM PLPEEVEES NNGPINNDYV HPGHQSPKQD HKNKNIYYPH YFLDSGSKVY REKDIITHEE FTEELLSGKI NCKLETLTPL I IPDTSDEN GLKLQGNKPG HKNYKFFNIN GELMIPGSEL RGMLRTHFEA LTKSCFAIFG EDSTLSWRMN ADEKDYKIDS NS IRKMESQ RNPKYRIPDE LQKELRNSGN GLFNRLYTSE RRFWSDVSNK FENSIDYKRE ILRCAGRPKN YKGGIIRQRK DSL MAEELK VHRLPLYDNF DIPDSAYKAN DHCRKSATCS TSRGCRERFT CGIKVRDKNR VFLNAANNNR QYLNNIKKSN HDLY LQYLK GEKKIRFNSK VITGSERSPI DVIAELNERG RQTGFIKLSG LNNSNKSQGN TGTTFNSGWD RFELNILLDD LETRP SKSD YPRPRLLFTK DQYEYNITKR CERVFEIDKG NKTGYPVDDQ IKKNYEDILD SYDGIKDQEV AERFDTFTRG SKLKVG DLV YFHIDGDNKI DSLIPVRISR KCASKTLGGK LDKALHPCTG LSDGLCPGCH LFGTTDYKGR VKFGFAKYEN GPEWLIT RG NNPERSLTLG VLESPRPAFS IPDDESEIPG RKFYLHHNGW RIIRQKQLEI RETVQPERNV TTEVMDKGNV FSFDVRFE N LREWELGLLL QSLDPGKNIA HKLGKGKPYG FGSVKIKIDS LHTFKINSNN DKIKRVPQSD IREYINKGYQ KLIEWSGNN SIQKGNVLPQ WHVIPHIDKL YKLLWVPFLN DSKLEPDVRY PVLNEESKGY IEGSDYTYKK LGDKDNLPYK TRVKGLTTPW SPWNPFQVI AEHEEQEVNV TGSRPSVTDK IERDGKMV UniProtKB: RAMP superfamily protein |
-Macromolecule #2: RNA (32-MER)
| Macromolecule | Name: RNA (32-MER) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Candidatus Scalindua brodae (bacteria) Candidatus Scalindua brodae (bacteria) |
| Molecular weight | Theoretical: 10.058966 KDa |
| Sequence | String: ACUUAAUGUC ACGGUACCCA AUUUUCUGCC CC |
-Macromolecule #3: RNA (5'-R(P*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP*GP*U)-3')
| Macromolecule | Name: RNA (5'-R(P*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP*GP*U)-3') type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Candidatus Scalindua brodae (bacteria) Candidatus Scalindua brodae (bacteria) |
| Molecular weight | Theoretical: 14.915912 KDa |
| Sequence | String: CUCUAGUAAC AGCCGUGGAG UCCGGGGCAG AAAAUUGGGU GAUUAA |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
| Details | No further treatment. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
 Movie
Movie Controller
Controller





 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)