+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | GTPgammaS Tube decorated with kinesin | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / PROTEIN FIBRIL | |||||||||
| Function / homology |  Function and homology information Function and homology informationHSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / COPI-mediated anterograde transport / COPI-independent Golgi-to-ER retrograde traffic / COPI-dependent Golgi-to-ER retrograde traffic / Kinesins / Neutrophil degranulation / astral microtubule / lysosome localization / structural constituent of cytoskeleton / microtubule cytoskeleton organization ...HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / COPI-mediated anterograde transport / COPI-independent Golgi-to-ER retrograde traffic / COPI-dependent Golgi-to-ER retrograde traffic / Kinesins / Neutrophil degranulation / astral microtubule / lysosome localization / structural constituent of cytoskeleton / microtubule cytoskeleton organization / spindle / mitotic cell cycle / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / microtubule / hydrolase activity / GTPase activity / centrosome / GTP binding / perinuclear region of cytoplasm / metal ion binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 6.8 Å | |||||||||
 Authors Authors | Zhou J / Wang H-W | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural insights into the mechanism of GTP initiation of microtubule assembly. Authors: Ju Zhou / Anhui Wang / Yinlong Song / Nan Liu / Jia Wang / Yan Li / Xin Liang / Guohui Li / Huiying Chu / Hong-Wei Wang /    Abstract: In eukaryotes, the dynamic assembly of microtubules (MT) plays an important role in numerous cellular processes. The underlying mechanism of GTP triggering MT assembly is still unknown. Here, we ...In eukaryotes, the dynamic assembly of microtubules (MT) plays an important role in numerous cellular processes. The underlying mechanism of GTP triggering MT assembly is still unknown. Here, we present cryo-EM structures of tubulin heterodimer at their GTP- and GDP-bound states, intermediate assembly states of GTP-tubulin, and final assembly stages of MT. Both GTP- and GDP-tubulin heterodimers adopt similar curved conformations with subtle flexibility differences. In head-to-tail oligomers of tubulin heterodimers, the inter-dimer interface of GDP-tubulin exhibits greater flexibility, particularly in tangential bending. Cryo-EM of the intermediate assembly states reveals two types of tubulin lateral contacts, "Tube-bond" and "MT-bond". Further, molecular dynamics (MD) simulations show that GTP triggers lateral contact formation in MT assembly in multiple sequential steps, gradually straightening the curved tubulin heterodimers. Therefore, we propose a flexible model of GTP-initiated MT assembly, including the formation of longitudinal and lateral contacts, to explain the nucleation and assembly of MT. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34080.map.gz emd_34080.map.gz | 171 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34080-v30.xml emd-34080-v30.xml emd-34080.xml emd-34080.xml | 18.4 KB 18.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34080_fsc.xml emd_34080_fsc.xml | 28.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_34080.png emd_34080.png | 96.7 KB | ||
| Filedesc metadata |  emd-34080.cif.gz emd-34080.cif.gz | 6 KB | ||
| Others |  emd_34080_half_map_1.map.gz emd_34080_half_map_1.map.gz emd_34080_half_map_2.map.gz emd_34080_half_map_2.map.gz | 1.5 GB 1.5 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34080 http://ftp.pdbj.org/pub/emdb/structures/EMD-34080 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34080 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34080 | HTTPS FTP |
-Related structure data
| Related structure data |  7ysqMC  7ysnC  7ysoC  7yspC  7ysrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34080.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34080.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.33 Å | ||||||||||||||||||||||||||||||||||||
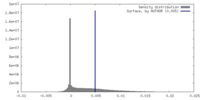
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34080_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
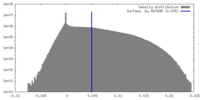
| Density Histograms |
-Half map: #2
| File | emd_34080_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : GTPgammaS-tube decorated with kinesin
| Entire | Name: GTPgammaS-tube decorated with kinesin |
|---|---|
| Components |
|
-Supramolecule #1: GTPgammaS-tube decorated with kinesin
| Supramolecule | Name: GTPgammaS-tube decorated with kinesin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 440 KDa |
-Macromolecule #1: Tubulin alpha chain
| Macromolecule | Name: Tubulin alpha chain / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.96125 KDa |
| Sequence | String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TVGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVVDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIVDLVL DRIRKLADQC TGLQGFLIFH SFGGGTGSGF TSLLMERLSV D YGKKSKLE ...String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TVGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVVDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIVDLVL DRIRKLADQC TGLQGFLIFH SFGGGTGSGF TSLLMERLSV D YGKKSKLE FAIYPAPQVS TAVVEPYNSI LTTHTTLEHS DCAFMVDNEA IYDICRRNLD IERPTYTNLN RLIGQIVSSI TA SLRFDGA LNVDLTEFQT NLVPYPRIHF PLVTYAPVIS AEKAYHEQLS VAEITNACFE PANQMVKCDP RHGKYMACCM LYR GDVVPK DVNAAIATIK TKRTIQFVDW CPTGFKVGIN YQPPTVVPGG DLAKVQRAVC MLSNTTAIAE AWARLDHKFD LMYA KRAFV HWYVGEGMEE GEFSEAREDL AALEKDYEEV GMDSGDGEGE GAEEY UniProtKB: Tubulin alpha-1 chain |
-Macromolecule #2: Tubulin beta-1 chain
| Macromolecule | Name: Tubulin beta-1 chain / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.194137 KDa |
| Sequence | String: MREIVHIQAG QCGNQIGAKF WEIISDEHGI DATGAYHGDS DLQLERINVY YNEASGGKYV PRAVLVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKEAESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTYS ...String: MREIVHIQAG QCGNQIGAKF WEIISDEHGI DATGAYHGDS DLQLERINVY YNEASGGKYV PRAVLVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKEAESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTYS VVPSPKVSDT VVEPYNATLS VHQLVENTDE TYCIDNEALY DICFRTLKLT TPTYGDLNHL VSLTMSGVTT CL RFPGQLN ADLRKLAVNM VPFPRLHFFM PGFAPLTSRG SQQYRALTVP ELTQQMFDAK NMMAACDPRH GRYLTVAAIF RGR MSMKEV DEQMLNIQNK NSSYFVEWIP NNVKTAVCDI PPRGLKMSAT FIGNSTAIQE LFKRISEQFT AMFRRKAFLH WYTG EGMDE MEFTEAESNM NDLVSEYQQY QEATADEDAE FEEEQEAEVD EN UniProtKB: Tubulin beta-1 chain |
-Macromolecule #3: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 4 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Macromolecule #4: 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE
| Macromolecule | Name: 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE / type: ligand / ID: 4 / Number of copies: 4 / Formula: GSP |
|---|---|
| Molecular weight | Theoretical: 539.246 Da |
| Chemical component information |  ChemComp-GSP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)