[English] 日本語
 Yorodumi
Yorodumi- EMDB-34076: GluK1-1a in nanodisc captured in SYM2081 bound desensitized state -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
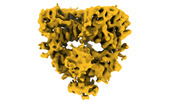
| Title | GluK1-1a in nanodisc captured in SYM2081 bound desensitized state | |||||||||
 Map data Map data | GluK1-1a nanodisc captured in desensitized state | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Kainate receptor / GluK1-1a splice variant / nanodisc / SYM2081 bound / desensitized state / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationligand-gated monoatomic ion channel activity / signaling receptor activity / postsynaptic membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.2 Å | |||||||||
 Authors Authors | Dhingra S / Kumar J | |||||||||
| Funding support |  India, 1 items India, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2023 Journal: Elife / Year: 2023Title: Functional Implications of the Exon 9 Splice Insert in GluK1 Kainate Receptors Authors: Dhingra S / Chopade PM / Vinnakota R / Kumar J | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34076.map.gz emd_34076.map.gz | 57.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34076-v30.xml emd-34076-v30.xml emd-34076.xml emd-34076.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34076.png emd_34076.png | 67.7 KB | ||
| Filedesc metadata |  emd-34076.cif.gz emd-34076.cif.gz | 7 KB | ||
| Others |  emd_34076_half_map_1.map.gz emd_34076_half_map_1.map.gz emd_34076_half_map_2.map.gz emd_34076_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34076 http://ftp.pdbj.org/pub/emdb/structures/EMD-34076 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34076 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34076 | HTTPS FTP |
-Validation report
| Summary document |  emd_34076_validation.pdf.gz emd_34076_validation.pdf.gz | 902.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34076_full_validation.pdf.gz emd_34076_full_validation.pdf.gz | 902.3 KB | Display | |
| Data in XML |  emd_34076_validation.xml.gz emd_34076_validation.xml.gz | 12.4 KB | Display | |
| Data in CIF |  emd_34076_validation.cif.gz emd_34076_validation.cif.gz | 14.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34076 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34076 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34076 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34076 | HTTPS FTP |
-Related structure data
| Related structure data |  7ysjMC  7ysvC  8gprC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34076.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34076.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GluK1-1a nanodisc captured in desensitized state | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.41 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: GluK1-1a nanodisc captured in desensitized state halfmap A
| File | emd_34076_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GluK1-1a nanodisc captured in desensitized state_halfmap_A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: GluK1-1a nanodisc captured in desensitized state halfmap B
| File | emd_34076_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GluK1-1a nanodisc captured in desensitized state_halfmap_B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of GluK1-1a in lipid nanodisc with SYM2081
| Entire | Name: Complex of GluK1-1a in lipid nanodisc with SYM2081 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of GluK1-1a in lipid nanodisc with SYM2081
| Supramolecule | Name: Complex of GluK1-1a in lipid nanodisc with SYM2081 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: EGFP is covalently conjugated to the C-terminus of GluK1-1a. MSP1E3D1 soybean polar lipid nanodisc used. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 600 KDa |
-Macromolecule #1: Glutamate receptor
| Macromolecule | Name: Glutamate receptor / type: protein_or_peptide / ID: 1 Details: L838-S843 correspond to Thrombin recognition site, A844-A847 correspond to Ala linker, V848-S1090 correspond to enhanced green fluorescent protein (EGFP), H1091-H1098 correspond to Octa-His affinity tag. Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 124.527125 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVLRIGGIFE TVENEPVNVE ELAFKFAVTS INRNRTLMPN TTLTYDIQRI NLFDSFEASR RACDQLALGV AALFGPSHSS SVSAVQSIC NALEVPHIQT RWKHPSVDSR DLFYINLYPD YAAISRAVLD LVLYYNWKTV TVVYEDSTGL IRLQELIKAP S RYNIKIKI ...String: QVLRIGGIFE TVENEPVNVE ELAFKFAVTS INRNRTLMPN TTLTYDIQRI NLFDSFEASR RACDQLALGV AALFGPSHSS SVSAVQSIC NALEVPHIQT RWKHPSVDSR DLFYINLYPD YAAISRAVLD LVLYYNWKTV TVVYEDSTGL IRLQELIKAP S RYNIKIKI RQLPPANKDA KPLLKEMKKS KEFYVIFDCS HETAAEILKQ ILFMGMMTEY YHYFFTTLDL FALDLELYRY SG VNMTGFR LLNIDNPHVS SIIEKWSMER LQAPPRPETG LLDGMMTTEA ALMYDAVYMV AIASHRASQL TVSSLQCHRH KPW RLGPRF MNLIKEARWD GLTGRITFNK TDGLRKDFDL DIISLKEEGT EKASGEVSKH LYKVWKKIGI WNSNSGLNMT DGNR DRSNN ITDSLANRTL IVTTILEEPY VMYRKSDKPL YGNDRFEGYC LDLLKELSNI LGFLYDVKLV PDGKYGAQND KGEWN GMVK ELIDHRADLA VAPLTITYVR EKVIDFSKPF MTLGISILYR KPNGTNPGVF SFLNPLSPDI WMYVLLAYLG VSVVLF VIA RFTPYEWYNP HPCNPDSDVV ENNFTLLNSF WFGVGALMQQ GSELMPKALS TRIVGGIWWF FTLIIISSYT ANLAAFL TV ERMESPIDSA DDLAKQTKIE YGAVRDGSTM TFFKKSKIST YEKMWAFMSS RQQSALVKNS DEGIQRVLTT DYALLMES T SIEYVTQRNC NLTQIGGLID SKGYGVGTPI GSPYRDKITI AILQLQEEGK LHMMKEKWWR GNGCPEEDSK EASALGVEN IGGIFIVLAA GLVLSVFVAI GEFLYKSRKN NDVEQHYLVP RGSAAAAVSK GEELFTGVVP ILVELDGDVN GHKFSVSGEG EGDATYGKL TLKFICTTGK LPVPWPTLVT TLTYGVQCFS RYPDHMKQHD FFKSAMPEGY VQERTIFFKD DGNYKTRAEV K FEGDTLVN RIELKGIDFK EDGNILGHKL EYNYNSHNVY IMADKQKNGI KVNFKIRHNI EDGSVQLADH YQQNTPIGDG PV LLPDNHY LSTQSKLSKD PNEKRDHMVL LEFVTAAGIT LGMDELYKSG LRSHHHHHHH H UniProtKB: Glutamate receptor |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.87 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average exposure time: 12.0 sec. / Average electron dose: 40.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)