[English] 日本語
 Yorodumi
Yorodumi- EMDB-33986: Cryo-EM structure of RNA polymerase in complex with P protein tet... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of RNA polymerase in complex with P protein tetramer of Newcastle disease virus | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cryo-EM / L-P complex / Newcastle disease virus / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationGDP polyribonucleotidyltransferase / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / Transferases; Transferring one-carbon groups; Methyltransferases / virion component / host cell cytoplasm / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / RNA-directed RNA polymerase / RNA-directed RNA polymerase activity / GTPase activity / ATP binding Similarity search - Function | |||||||||
| Biological species |  Avian orthoavulavirus 1 Avian orthoavulavirus 1 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Chen Y / Jingyuan C | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure of the Newcastle Disease Virus L protein in complex with tetrameric phosphoprotein. Authors: Jingyuan Cong / Xiaoying Feng / Huiling Kang / Wangjun Fu / Lei Wang / Chenlong Wang / Xuemei Li / Yutao Chen / Zihe Rao /  Abstract: Newcastle disease virus (NDV) belongs to Paramyxoviridae, which contains lethal human and animal pathogens. NDV RNA genome is replicated and transcribed by a multifunctional 250 kDa RNA-dependent ...Newcastle disease virus (NDV) belongs to Paramyxoviridae, which contains lethal human and animal pathogens. NDV RNA genome is replicated and transcribed by a multifunctional 250 kDa RNA-dependent RNA polymerase (L protein). To date, high-resolution structure of NDV L protein complexed with P protein remains to be elucidated, limiting our understanding of the molecular mechanisms of Paramyxoviridae replication/transcription. Here, we used cryo-EM and enzymatic assays to investigate the structure-function relationship of L-P complex. We found that C-terminal of CD-MTase-CTD module of the atomic-resolution L-P complex conformationally rearranges, and the priming/intrusion loops are likely in RNA elongation conformations different from previous structures. The P protein adopts a unique tetrameric organization and interacts with L protein. Our findings indicate that NDV L-P complex represents elongation state distinct from previous structures. Our work greatly advances the understanding of Paramyxoviridae RNA synthesis, revealing how initiation/elongation alternates, providing clues for identifying therapeutic targets against Paramyxoviridae. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33986.map.gz emd_33986.map.gz | 28.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33986-v30.xml emd-33986-v30.xml emd-33986.xml emd-33986.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33986.png emd_33986.png | 20.3 KB | ||
| Filedesc metadata |  emd-33986.cif.gz emd-33986.cif.gz | 7.5 KB | ||
| Others |  emd_33986_half_map_1.map.gz emd_33986_half_map_1.map.gz emd_33986_half_map_2.map.gz emd_33986_half_map_2.map.gz | 15.6 MB 15.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33986 http://ftp.pdbj.org/pub/emdb/structures/EMD-33986 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33986 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33986 | HTTPS FTP |
-Related structure data
| Related structure data |  7yotMC  7youC  7yovC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33986.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33986.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
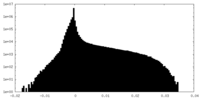
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_33986_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
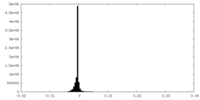
| Density Histograms |
-Half map: #2
| File | emd_33986_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
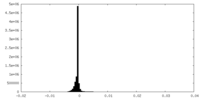
| Density Histograms |
- Sample components
Sample components
-Entire : NDV L-P complex
| Entire | Name: NDV L-P complex |
|---|---|
| Components |
|
-Supramolecule #1: NDV L-P complex
| Supramolecule | Name: NDV L-P complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Avian orthoavulavirus 1 Avian orthoavulavirus 1 |
-Macromolecule #1: NDV P protein
| Macromolecule | Name: NDV P protein / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Avian orthoavulavirus 1 Avian orthoavulavirus 1 |
| Molecular weight | Theoretical: 41.738379 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MATFTDAEID ELFETSGTVI DSIITAQGKP VETVGRSAIP QGKTKALSLA WEKHGNTNTP AAQESAGEQD QHGQNQASNS NRATPEEGP HSSQAQAATQ PQEDANESQL KTGASSSLLS MLDKLSNKSS NAKKGPPQSP PQQALHSKGS PAVEQTQHGA N QGRAQQET ...String: MATFTDAEID ELFETSGTVI DSIITAQGKP VETVGRSAIP QGKTKALSLA WEKHGNTNTP AAQESAGEQD QHGQNQASNS NRATPEEGP HSSQAQAATQ PQEDANESQL KTGASSSLLS MLDKLSNKSS NAKKGPPQSP PQQALHSKGS PAVEQTQHGA N QGRAQQET GHQAAPSPGP PGTGVNIAFP GQRGVSPQSV GATQPAPQSG QNQGSTPASA DHVQPPVDFV QAMMSMMEAI SQ RVSKIDY QLDLVLKQTS SIPTMRSEIQ QLKTSVAVME ANLGMMKILD PGCANVSSLS DLRAVAKSHP VLIAGPGDPS PYV TQGGEI ALNKLSQPVP HPSDLIKHAT SGGPDIGIER DTVRALILSR PMHPSSSSKL LSKLDSAGSV EEIRKIKRLA LNG UniProtKB: Phosphoprotein |
-Macromolecule #2: RNA-directed RNA polymerase L
| Macromolecule | Name: RNA-directed RNA polymerase L / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Avian orthoavulavirus 1 Avian orthoavulavirus 1 |
| Molecular weight | Theoretical: 249.183188 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAGSGSERAE HQIILPESHL SSPLVKHKLL YYWKLTGLPL PDECDFDHLI LSRQWKKILE SSTPDIERMI KLGRSVHQTL SHSSKLTGI LHPRCLEDLV GLDIPDSTNK FRRIEKKIQI HNTRYGEPFT RLCSYVEKKL LGSSWTHKIR RSEEFDSLRT D PAFWFHSS ...String: MAGSGSERAE HQIILPESHL SSPLVKHKLL YYWKLTGLPL PDECDFDHLI LSRQWKKILE SSTPDIERMI KLGRSVHQTL SHSSKLTGI LHPRCLEDLV GLDIPDSTNK FRRIEKKIQI HNTRYGEPFT RLCSYVEKKL LGSSWTHKIR RSEEFDSLRT D PAFWFHSS WSTAKFAWLH VKQIQRHLIV AARTRSASNK LVTLSHRSGQ VFITPELVIV THTNENKFTC LSQELVLMYA DM MEGRDMV NIISSTAVHL RCLAEKIDDI LRLVDALARD LGNQVYDVVA LMEGFAYGAV QLLEPSGTFA GDFFSFNLQE LRD TLICLL PQRIADSVTH AIANIFSGLE QNQAAEMLCL LRLWGHPLLE SRAAAKAVRA QMCAPKMVDF DMILQVLSFF KGTI INGYR KKNAGVWPRV KAHTIYGNVI AQLHADSAEI SHDIMLREYK NLSAIEFEAC IEYDPVTNLS MFLKDKAIAH PRNNW LASF RRNLLSEEQK KNVQDSTSTN RLLIEFLESN DFDPYKEMEY LTTLEYLRDD SVAVSYSLKE EEVKVNGRIF AKLTKK LRN CQVMAEGILA DQIAPFFQGN GVIQDSISLT KSMLAMSQLS YNSNRKRITD CKERVSSSRN HDLKGKHRRR VATFITT DL QKYCLNWRYQ TIKLFAHAIN QLMGLPHFFE WIHLRLMDTT MFVGDPFNPP SDPTDYDLTK VPNDDIYIVS ARGGIEGL C QKLWTMISIA AIQLAAARSH CRVACMVQGD NQVIAVTREV RPDDSPESVL TQLHEASDNF FRELIHVNHL IGHNLKDRE TIRSDTFFIY SKRIFKDGAI LSQVLKNSSK LVLVSGDLSE NTVMSCANIS STVARLCENG LPKDFCYYLN YLMSCIQTYF DSEFSITSS TQSGSNQSWI NDIPFIHSYV LTPAQLGGLS NLQYSRLYTR NIGDPGTTAF AEVKRLEAVG LLGPNIMTNI L TRPPGNGD WASLCNDPYS FNFESVASPS IVLKKHTQRV LFETCSNPLL SGVHTEDNEA EEKALAEYLL NQEVIHPRVA HA IMEASSV GRRKQIQGLV DTTNTVIKIA LSRKPLGIKR LARIINYSSM HAMLFRDDVF LSNRANHPLV SSDMCSLALA DYA RNRSWS PLTGGRKILG VSNPDTIELV EGEILSISGG CSKCDSGDEQ FTWFHLPSNI ELTDDTSKNP PMRVPYLGSK TQER RAASL AKIAHMSPHV KAALRASSVL IWAYGDNDIN WTAALKLARS RCNISSEYLR LLSPLPTAGN LQHRLDDGIT QMTFT PASL YRVSPYVHIS NDSQRLFTEE GVKEGNVVYQ QIMLLGLSLI ESLFPMTVTK TYDEITLHLH SKFSCCIREA PVAVPF ELT GVAPDLRVVA SNKFMYDPNP VAEGDFARLD LAIFKSYELN LESYSTVELM NILSISSGKL IGQSVVSYDE ETSIKND AI IVYDNTRNWI SEAQNSDVVR LFEYAALEVL LDCSYQLYYL RVRGLNNVVL YMSDLYKNMP GILLSNIAAT ISHPIIHS R LHTVGLISHD GSHQLADTDF IELSAKLLVS CTRRVVSGLY AGNKYDLLFP SVLDDNLNEK MLQLISRLCC LYTVLFATT REIPKIRGLP AEEKCAMLTE YLLSDAVRPL LSPEQVDSIT SPSIVTFPAN LYYMSRKSLN LIREREDRDS ILALMFPQEP LFEFPLVQD IGARVKDQLT MKPAAFLHEL DLSAPARYDA YTLEQARSDC ALADMGEDQL VRYLFRGVGT ASSSWYKASH L LSVPEIRC ARHGNSLYLA EGSGAIMSLL ELHIPHETIY YNTLFSNEMN PPQRHFGPTP TQFLNSVVYR NLQAEVPCKD GF VQEFRTL WRENTEESDL TSDKAVGYIT SVVPYRSVSL LHCDIEIPPG SNQSLLDQLA TNLSLIAMHS VREGGVVIVK ILY SMGYYF HLLVNLFTPC SVKGYVLSNG YACRGDMECY VVFVMGYLGG PTFVNEVVRM AKTLIQRHGT LLAKSDETAL MALF TSQKQ RVDNILSSPL PRLAKLLRRN IDTALIEAGG QPVRPFCAES LVNTLSDITQ TTQVIASHID TVIRSVIYME AEGDL ADTV FLFTPYNLSI DGKKRTSLKQ CTRQILEVTI LGLGPEDLNR VGDIISLILR GTISLEDLIP LRTYLKMSTC PKYLKS VLG LTKLREMFSD GSMLYLTRAQ QKFYMKTVGN AVKGYYNSSK NENLYFQG UniProtKB: RNA-directed RNA polymerase L |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)