[English] 日本語
 Yorodumi
Yorodumi- EMDB-33928: Cryo-EM structure of Nb29-alpha1AAR-miniGsq complex bound to nora... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Nb29-alpha1AAR-miniGsq complex bound to noradrenaline | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / Nanobody / Agonist / Complex / MEMBRANE PROTEIN | |||||||||
| Biological species | synthetic construct (others) /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.52 Å | |||||||||
 Authors Authors | Toyoda Y / Zhu A / Yan C / Kobilka BK / Liu X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis of α-adrenergic receptor activation and recognition by an extracellular nanobody. Authors: Yosuke Toyoda / Angqi Zhu / Fang Kong / Sisi Shan / Jiawei Zhao / Nan Wang / Xiaoou Sun / Linqi Zhang / Chuangye Yan / Brian K Kobilka / Xiangyu Liu /    Abstract: The αadrenergic receptor (αAR) belongs to the family of G protein-coupled receptors that respond to adrenaline and noradrenaline. αAR is involved in smooth muscle contraction and cognitive ...The αadrenergic receptor (αAR) belongs to the family of G protein-coupled receptors that respond to adrenaline and noradrenaline. αAR is involved in smooth muscle contraction and cognitive function. Here, we present three cryo-electron microscopy structures of human αAR bound to the endogenous agonist noradrenaline, its selective agonist oxymetazoline, and the antagonist tamsulosin, with resolutions range from 2.9 Å to 3.5 Å. Our active and inactive αAR structures reveal the activation mechanism and distinct ligand binding modes for noradrenaline compared with other adrenergic receptor subtypes. In addition, we identified a nanobody that preferentially binds to the extracellular vestibule of αAR when bound to the selective agonist oxymetazoline. These results should facilitate the design of more selective therapeutic drugs targeting both orthosteric and allosteric sites in this receptor family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33928.map.gz emd_33928.map.gz | 40.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33928-v30.xml emd-33928-v30.xml emd-33928.xml emd-33928.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33928.png emd_33928.png | 50.5 KB | ||
| Filedesc metadata |  emd-33928.cif.gz emd-33928.cif.gz | 6.3 KB | ||
| Others |  emd_33928_half_map_1.map.gz emd_33928_half_map_1.map.gz emd_33928_half_map_2.map.gz emd_33928_half_map_2.map.gz | 39.8 MB 39.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33928 http://ftp.pdbj.org/pub/emdb/structures/EMD-33928 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33928 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33928 | HTTPS FTP |
-Related structure data
| Related structure data |  7ymhMC  7ym8C  7ymjC C: citing same article ( M: atomic model generated by this map |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33928.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33928.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0979 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33928_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
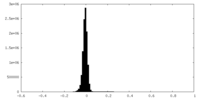
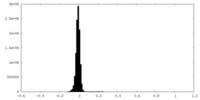
| Density Histograms |
-Half map: #1
| File | emd_33928_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
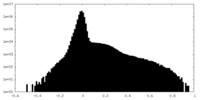
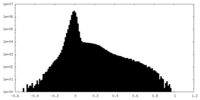
| Density Histograms |
- Sample components
Sample components
-Entire : Nb29-alpha1AAR-miniGsq complex
| Entire | Name: Nb29-alpha1AAR-miniGsq complex |
|---|---|
| Components |
|
-Supramolecule #1: Nb29-alpha1AAR-miniGsq complex
| Supramolecule | Name: Nb29-alpha1AAR-miniGsq complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Supramolecule #2: Nb29
| Supramolecule | Name: Nb29 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Supramolecule #3: alpha1AAR
| Supramolecule | Name: alpha1AAR / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #4: miniGsq
| Supramolecule | Name: miniGsq / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: alpha1A-adrenergic receptor
| Macromolecule | Name: alpha1A-adrenergic receptor / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 53.736715 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDAMVFLSG QASDSSQCTQ PPAPVQISKA ILLGVILGGL ILFGVLGNIL VILSVACHRH LHSVTHYYI VNLAVADLLL TSTVLPFSAI FEVLGYWAFG RVFCNIWAAV DVLCCTASIM GLCIISIDRY IGVSYPLRYP T IVTQRRGL ...String: MKTIIALSYI FCLVFADYKD DDDAMVFLSG QASDSSQCTQ PPAPVQISKA ILLGVILGGL ILFGVLGNIL VILSVACHRH LHSVTHYYI VNLAVADLLL TSTVLPFSAI FEVLGYWAFG RVFCNIWAAV DVLCCTASIM GLCIISIDRY IGVSYPLRYP T IVTQRRGL MALLCVWALS LVISIGPLFG WRQPAPEDET ICQINEEPGY VLFSALGSFY LPLAIILVMY CRVYVVAKRE SR GLKSGLN IFEMLRIDEG GGSGGDEAEK LFNQDVDAAV RGILRNAKLK PVYDSLDAVR RAALINMVFQ MGETGVAGFT NSL RMLQQK RWDEAAVNLA KSRWYNQTPN RAKRVITTFR TGTWDAYLKF SREKKAAKTL GIVVGCFVLC WLPFFLVMPI GSFF PDFKP SETVFKIVFW LGYLNSCINP IIYPCSSQEF KKAFQNVLRI QCLCRKQSSK HALGYTLHPP SQAVEGQHHH HHHHH |
-Macromolecule #2: miniGsq
| Macromolecule | Name: miniGsq / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.571365 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHHH LEVLFQGPIE KQLQKDKQVY RATHRLLLLG ADNSGKSTIV KQMRILHGGS GGSGGTSGIF ETKFQVDKVN FHMFDVGGQ RDERRKWIQC FNDVTAIIFV VDSSDYNRLQ EALNDFKSIW NNRWLRTISV ILFLNKQDLL AEKVLAGKSK I EDYFPEFA ...String: MGHHHHHHHH LEVLFQGPIE KQLQKDKQVY RATHRLLLLG ADNSGKSTIV KQMRILHGGS GGSGGTSGIF ETKFQVDKVN FHMFDVGGQ RDERRKWIQC FNDVTAIIFV VDSSDYNRLQ EALNDFKSIW NNRWLRTISV ILFLNKQDLL AEKVLAGKSK I EDYFPEFA RYTTPEDATP EPGEDPRVTR AKYFIRDEFL RISTASGDGR HYCYPHFTCA VDTENARRIF NDCKDIILQM NL REYNLV |
-Macromolecule #3: Nb29
| Macromolecule | Name: Nb29 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 15.815713 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKYLLPTAAA GLLLLAAQPA MAQVQLQESG GGLVQAGGSL RLSCAASGNI SAHWKMGWYR QAPGKEREFV AGIGYANTNY ADSVKGRFT ISRDNAKNTV YLQMNSLKPE DTAVYYCAAY SYYRDHSYWG QGTQVTVSSH HHHHH |
-Macromolecule #4: Noradrenaline
| Macromolecule | Name: Noradrenaline / type: ligand / ID: 4 / Number of copies: 1 / Formula: E5E |
|---|---|
| Molecular weight | Theoretical: 170.186 Da |
| Chemical component information |  ChemComp-E5E: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.3 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)