+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-3355 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Activation of NMDA receptors and the mechanism of inhibition by ifenprodil - Class X | |||||||||
 マップデータ マップデータ | Class X, unsharpened, unmasked map | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | NMDA receptor / glutamate receptor / GluN1 / GluN2B / ion channel | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報pons maturation / positive regulation of Schwann cell migration / regulation of cell communication / EPHB-mediated forward signaling / Assembly and cell surface presentation of NMDA receptors / olfactory learning / dendritic branch / conditioned taste aversion / regulation of respiratory gaseous exchange / protein localization to postsynaptic membrane ...pons maturation / positive regulation of Schwann cell migration / regulation of cell communication / EPHB-mediated forward signaling / Assembly and cell surface presentation of NMDA receptors / olfactory learning / dendritic branch / conditioned taste aversion / regulation of respiratory gaseous exchange / protein localization to postsynaptic membrane / propylene metabolic process / response to glycine / regulation of monoatomic cation transmembrane transport / voltage-gated monoatomic cation channel activity / Synaptic adhesion-like molecules / NMDA glutamate receptor activity / RAF/MAP kinase cascade / parallel fiber to Purkinje cell synapse / NMDA selective glutamate receptor complex / response to morphine / calcium ion transmembrane import into cytosol / glutamate binding / neuromuscular process / protein heterotetramerization / regulation of synapse assembly / positive regulation of calcium ion transport into cytosol / positive regulation of reactive oxygen species biosynthetic process / glycine binding / male mating behavior / regulation of axonogenesis / regulation of dendrite morphogenesis / suckling behavior / startle response / response to amine / monoatomic cation transmembrane transport / regulation of neuronal synaptic plasticity / associative learning / monoatomic cation transport / social behavior / excitatory synapse / ligand-gated monoatomic ion channel activity / positive regulation of excitatory postsynaptic potential / cellular response to glycine / positive regulation of dendritic spine maintenance / Unblocking of NMDA receptors, glutamate binding and activation / cellular response to manganese ion / phosphatase binding / glutamate receptor binding / long-term memory / regulation of neuron apoptotic process / prepulse inhibition / monoatomic cation channel activity / calcium ion homeostasis / glutamate-gated receptor activity / synaptic cleft / response to fungicide / glutamate-gated calcium ion channel activity / presynaptic active zone membrane / dendrite membrane / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / sensory perception of pain / response to amphetamine / ionotropic glutamate receptor signaling pathway / hippocampal mossy fiber to CA3 synapse / positive regulation of synaptic transmission, glutamatergic / regulation of membrane potential / adult locomotory behavior / excitatory postsynaptic potential / learning / synaptic transmission, glutamatergic / synaptic membrane / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / postsynaptic density membrane / regulation of long-term neuronal synaptic plasticity / visual learning / calcium channel activity / regulation of synaptic plasticity / terminal bouton / response to organic cyclic compound / cerebral cortex development / memory / synaptic vesicle membrane / response to calcium ion / intracellular calcium ion homeostasis / neuron cellular homeostasis / calcium ion transport / rhythmic process / synaptic vesicle / signaling receptor activity / presynaptic membrane / amyloid-beta binding / protein-containing complex assembly / chemical synaptic transmission / postsynaptic membrane / response to ethanol / negative regulation of neuron apoptotic process / transcription by RNA polymerase II / dendritic spine / postsynaptic density / learning or memory 類似検索 - 分子機能 | |||||||||
| 生物種 |  | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 6.25 Å | |||||||||
 データ登録者 データ登録者 | Tajima N / Karakas E / Grant T / Simorowski N / Diaz-Avalos R / Grigorieff N / Furukawa H | |||||||||
 引用 引用 |  ジャーナル: Nature / 年: 2016 ジャーナル: Nature / 年: 2016タイトル: Activation of NMDA receptors and the mechanism of inhibition by ifenprodil. 著者: Nami Tajima / Erkan Karakas / Timothy Grant / Noriko Simorowski / Ruben Diaz-Avalos / Nikolaus Grigorieff / Hiro Furukawa /  要旨: The physiology of N-methyl-d-aspartate (NMDA) receptors is fundamental to brain development and function. NMDA receptors are ionotropic glutamate receptors that function as heterotetramers composed ...The physiology of N-methyl-d-aspartate (NMDA) receptors is fundamental to brain development and function. NMDA receptors are ionotropic glutamate receptors that function as heterotetramers composed mainly of GluN1 and GluN2 subunits. Activation of NMDA receptors requires binding of neurotransmitter agonists to a ligand-binding domain (LBD) and structural rearrangement of an amino-terminal domain (ATD). Recent crystal structures of GluN1-GluN2B NMDA receptors bound to agonists and an allosteric inhibitor, ifenprodil, represent the allosterically inhibited state. However, how the ATD and LBD move to activate the NMDA receptor ion channel remains unclear. Here we applied X-ray crystallography, single-particle electron cryomicroscopy and electrophysiology to rat NMDA receptors to show that, in the absence of ifenprodil, the bi-lobed structure of GluN2 ATD adopts an open conformation accompanied by rearrangement of the GluN1-GluN2 ATD heterodimeric interface, altering subunit orientation in the ATD and LBD and forming an active receptor conformation that gates the ion channel. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_3355.map.gz emd_3355.map.gz | 19.8 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-3355-v30.xml emd-3355-v30.xml emd-3355.xml emd-3355.xml | 11.8 KB 11.8 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_3355.png emd_3355.png | 434.4 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3355 http://ftp.pdbj.org/pub/emdb/structures/EMD-3355 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3355 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3355 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_3355_validation.pdf.gz emd_3355_validation.pdf.gz | 242.3 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_3355_full_validation.pdf.gz emd_3355_full_validation.pdf.gz | 241.4 KB | 表示 | |
| XML形式データ |  emd_3355_validation.xml.gz emd_3355_validation.xml.gz | 6.2 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3355 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3355 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3355 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3355 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  5fxjMC  3352C  3353C  3354C  3356C  5b3jC  5fxgC  5fxhC  5fxiC  5fxkC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_3355.map.gz / 形式: CCP4 / 大きさ: 62.5 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_3355.map.gz / 形式: CCP4 / 大きさ: 62.5 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Class X, unsharpened, unmasked map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
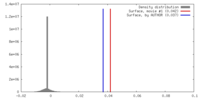
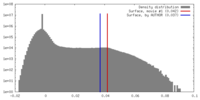
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : NMDA Receptor
| 全体 | 名称: NMDA Receptor |
|---|---|
| 要素 |
|
-超分子 #1000: NMDA Receptor
| 超分子 | 名称: NMDA Receptor / タイプ: sample / ID: 1000 詳細: The sample was purified in the presence of agonists Glycine and L-glutamate. 集合状態: One heterotetramer of 2 GluN1 and 2 GluN2B subunits Number unique components: 2 |
|---|---|
| 分子量 | 理論値: 370 KDa |
-分子 #1: N-methyl-D-aspartate receptor GluN1
| 分子 | 名称: N-methyl-D-aspartate receptor GluN1 / タイプ: protein_or_peptide / ID: 1 / Name.synonym: GluN1, NR1 / コピー数: 2 / 集合状態: dimer / 組換発現: Yes |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 93 KDa |
| 組換発現 | 生物種:  組換株: Sf9 CRL-1711 / 組換細胞: Sf9 / 組換プラスミド: Modified pFL and pUCDM |
| 配列 | UniProtKB: Glutamate receptor ionotropic, NMDA 1 |
-分子 #2: N-methyl-D-aspartate receptor GluN2B
| 分子 | 名称: N-methyl-D-aspartate receptor GluN2B / タイプ: protein_or_peptide / ID: 2 / Name.synonym: GluN2B, NR2B / コピー数: 2 / 集合状態: Dimer / 組換発現: Yes |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 92 KDa |
| 組換発現 | 生物種:  組換株: Sf9 CRL-1711 / 組換細胞: Sf9 / 組換プラスミド: Modified pFL and pUCDM |
| 配列 | UniProtKB: Gliding motility protein GldL |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 2 mg/mL |
|---|---|
| 緩衝液 | pH: 7 詳細: 200 mM NaCl, 20 mM HEPES pH 7.0, 10 mM Glycine, 10 mM L-Glutamate, 0.002% MNG-3 |
| グリッド | 詳細: C-flat 1.2/1.3 Cu 400 |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 90 % / 装置: FEI VITROBOT MARK II / 手法: 3s Blot time |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| アライメント法 | Legacy - Electron beam tilt params: 0 |
| 詳細 | 21s exposure into 70 frames, with an exposure rate of ~8 electrons/pixel/s on the camera. |
| 日付 | 2015年8月10日 |
| 撮影 | カテゴリ: CCD フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 実像数: 1200 / 平均電子線量: 100 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 倍率(補正後): 38168 / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): 2.5 µm / 最小 デフォーカス(公称値): 1.0 µm / 倍率(公称値): 22500 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
- 画像解析
画像解析
| CTF補正 | 詳細: Each Particle |
|---|---|
| 最終 再構成 | 想定した対称性 - 点群: C2 (2回回転対称) / アルゴリズム: OTHER / 解像度のタイプ: BY AUTHOR / 解像度: 6.25 Å / 解像度の算出法: OTHER / ソフトウェア - 名称: Unblur, CTFFIND4, FREALIGN 詳細: The highest resolution included in the refinement was 8A. 使用した粒子像数: 14000 |
-原子モデル構築 1
| 初期モデル | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B / Chain - #2 - Chain ID: C / Chain - #3 - Chain ID: D |
|---|---|
| ソフトウェア | 名称:  Coot Coot |
| 詳細 | The individual domains were initially fitted using coot and real space refinement was performed using Phenix |
| 精密化 | 空間: REAL / プロトコル: FLEXIBLE FIT / 当てはまり具合の基準: Real Space |
| 得られたモデル |  PDB-5fxj: |
 ムービー
ムービー コントローラー
コントローラー













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)