+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of PEIP-Bs_enolase complex | |||||||||
 Map data Map data | Cryo-EM structure of PEIP-Bs_enolase complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Enolase inhibitor / Glycolysis / Bacteriophage / ANTIMICROBIAL PROTEIN / LYASE-LYASE INHIBITOR complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationphosphopyruvate hydratase / phosphopyruvate hydratase complex / phosphopyruvate hydratase activity / sporulation resulting in formation of a cellular spore / glycolytic process / cell surface / magnesium ion binding / extracellular region Similarity search - Function | |||||||||
| Biological species |   Bacillus phage SP01 (virus) Bacillus phage SP01 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Li S / Zhang K | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2022 Journal: Cell Rep / Year: 2022Title: Bacteriophage protein PEIP is a potent Bacillus subtilis enolase inhibitor. Authors: Kaining Zhang / Shanshan Li / Yawen Wang / Zhihao Wang / Nancy Mulvenna / Hang Yang / Peipei Zhang / Huan Chen / Yan Li / Hongliang Wang / Yongxiang Gao / Sivaramesh Wigneshweraraj / Steve ...Authors: Kaining Zhang / Shanshan Li / Yawen Wang / Zhihao Wang / Nancy Mulvenna / Hang Yang / Peipei Zhang / Huan Chen / Yan Li / Hongliang Wang / Yongxiang Gao / Sivaramesh Wigneshweraraj / Steve Matthews / Kaiming Zhang / Bing Liu /   Abstract: Enolase is a highly conserved enzyme that presents in all organisms capable of glycolysis or fermentation. Its immediate product phosphoenolpyruvate is essential for other important processes like ...Enolase is a highly conserved enzyme that presents in all organisms capable of glycolysis or fermentation. Its immediate product phosphoenolpyruvate is essential for other important processes like peptidoglycan synthesis and the phosphotransferase system in bacteria. Therefore, enolase inhibitors are of great interest. Here, we report that Gp60, a phage-encoded enolase inhibitor protein (PEIP) of bacteriophage SPO1 for Bacillus subtilis, is an enolase inhibitor. PEIP-expressing bacteria exhibit growth attenuation, thinner cell walls, and safranin color in Gram staining owing to impaired peptidoglycan synthesis. We solve the structure of PEIP-enolase tetramer and show that PEIP disassembles enolase by disrupting the basic dimer unit. The structure reveals that PEIP does not compete for substrate binding but induces a cascade of conformational changes that limit accessibility to the enolase catalytic site. This phage-inspired disassembly of enolase represents an alternative strategy for the development of anti-microbial drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33300.map.gz emd_33300.map.gz | 32.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33300-v30.xml emd-33300-v30.xml emd-33300.xml emd-33300.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33300.png emd_33300.png | 93.2 KB | ||
| Filedesc metadata |  emd-33300.cif.gz emd-33300.cif.gz | 5.9 KB | ||
| Others |  emd_33300_half_map_1.map.gz emd_33300_half_map_1.map.gz emd_33300_half_map_2.map.gz emd_33300_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33300 http://ftp.pdbj.org/pub/emdb/structures/EMD-33300 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33300 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33300 | HTTPS FTP |
-Related structure data
| Related structure data |  7xmlMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33300.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33300.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of PEIP-Bs_enolase complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map-2
| File | emd_33300_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map-2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
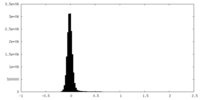
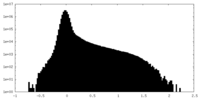
| Density Histograms |
-Half map: half map-1
| File | emd_33300_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map-1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
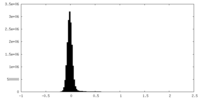
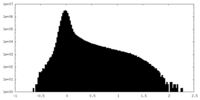
| Density Histograms |
- Sample components
Sample components
-Entire : PEIP-Bs_enolase complex
| Entire | Name: PEIP-Bs_enolase complex |
|---|---|
| Components |
|
-Supramolecule #1: PEIP-Bs_enolase complex
| Supramolecule | Name: PEIP-Bs_enolase complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Molecular weight | Theoretical: 100 KDa |
-Supramolecule #2: Bs_enolase
| Supramolecule | Name: Bs_enolase / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: PEIP
| Supramolecule | Name: PEIP / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Bacillus phage SP01 (virus) Bacillus phage SP01 (virus) |
-Macromolecule #1: Enolase
| Macromolecule | Name: Enolase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: phosphopyruvate hydratase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 46.626148 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPYIVDVYAR EVLDSRGNPT VEVEVYTETG AFGRALVPSG ASTGEYEAVE LRDGDKDRYL GKGVLTAVNN VNEIIAPELL GFDVTEQNA IDQLLIELDG TENKGKLGAN AILGVSMACA RAAADFLQIP LYQYLGGFNS KTLPVPMMNI VNGGEHADNN V DIQEFMIM ...String: MPYIVDVYAR EVLDSRGNPT VEVEVYTETG AFGRALVPSG ASTGEYEAVE LRDGDKDRYL GKGVLTAVNN VNEIIAPELL GFDVTEQNA IDQLLIELDG TENKGKLGAN AILGVSMACA RAAADFLQIP LYQYLGGFNS KTLPVPMMNI VNGGEHADNN V DIQEFMIM PVGAPNFREA LRMGAQIFHS LKSVLSAKGL NTAVGDEGGF APNLGSNEEA LQTIVEAIEK AGFKPGEEVK LA MDAASSE FYNKEDGKYH LSGEGVVKTS AEMVDWYEEL VSKYPIISIE DGLDENDWEG HKLLTERLGK KVQLVGDDLF VTN TKKLSE GIKNGVGNSI LIKVNQIGTL TETFDAIEMA KRAGYTAVIS HRSGETEDST IADIAVATNA GQIKTGAPSR TDRV AKYNQ LLRIEDQLAE TAQYHGINSF YNLNK UniProtKB: Enolase |
-Macromolecule #2: Putative gene 60 protein
| Macromolecule | Name: Putative gene 60 protein / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacillus phage SP01 (virus) Bacillus phage SP01 (virus) |
| Molecular weight | Theoretical: 8.558438 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLNQVEVLRE EYVEGYVVQM WRRNPSNAPV IEVFTEDNLE EGIIPEYVTA NDDTFDRIVD AVEFGYLEEL ELV UniProtKB: Putative gene 60 protein |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)