[English] 日本語
 Yorodumi
Yorodumi- EMDB-33123: Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with S309 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with S309 fab (local refinement) | |||||||||
 Map data Map data | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with S309 fab (local refinement) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / viral translation / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / viral translation / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / membrane fusion / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Gao GF / Qi JX / Zhao ZN / Xie YF / Liu S | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape Authors: Zhao Z / Zhou J / Tian M / Huang M / Liu S / Xie Y / Han P / Bai C / Han P / Zheng A / Fu L / Gao Y / Peng Q / Li Y / Chai Y / Zhang Z / Zhao X / Song H / Qi J / Wang Q / Wang P / Gao GF | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33123.map.gz emd_33123.map.gz | 482.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33123-v30.xml emd-33123-v30.xml emd-33123.xml emd-33123.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33123.png emd_33123.png | 75.8 KB | ||
| Filedesc metadata |  emd-33123.cif.gz emd-33123.cif.gz | 6.3 KB | ||
| Others |  emd_33123_half_map_1.map.gz emd_33123_half_map_1.map.gz emd_33123_half_map_2.map.gz emd_33123_half_map_2.map.gz | 474.5 MB 474.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33123 http://ftp.pdbj.org/pub/emdb/structures/EMD-33123 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33123 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33123 | HTTPS FTP |
-Validation report
| Summary document |  emd_33123_validation.pdf.gz emd_33123_validation.pdf.gz | 895.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33123_full_validation.pdf.gz emd_33123_full_validation.pdf.gz | 895.5 KB | Display | |
| Data in XML |  emd_33123_validation.xml.gz emd_33123_validation.xml.gz | 18.6 KB | Display | |
| Data in CIF |  emd_33123_validation.cif.gz emd_33123_validation.cif.gz | 22.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33123 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33123 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33123 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33123 | HTTPS FTP |
-Related structure data
| Related structure data |  7xckMC  7xchC  7xciC  7xcoC  7xcpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33123.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33123.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with S309 fab (local refinement) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.67 Å | ||||||||||||||||||||||||||||||||||||
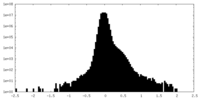
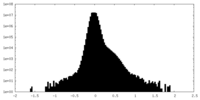
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex...
| File | emd_33123_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with S309 fab (local refinement) | ||||||||||||
| Projections & Slices |
| ||||||||||||
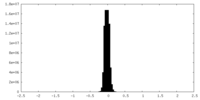
| Density Histograms |
-Half map: Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex...
| File | emd_33123_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with S309 fab (local refinement) | ||||||||||||
| Projections & Slices |
| ||||||||||||
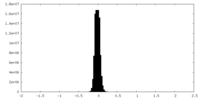
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with S309 ...
| Entire | Name: Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with S309 fab (local refinement) |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with S309 ...
| Supramolecule | Name: Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with S309 fab (local refinement) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: S309 fab
| Supramolecule | Name: S309 fab / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: SARS-CoV-2 Omicron RBD
| Supramolecule | Name: SARS-CoV-2 Omicron RBD / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|
-Macromolecule #1: S309 heavy chain
| Macromolecule | Name: S309 heavy chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.573471 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE VKKPGASVKV SCKASGYPFT SYGISWVRQA PGQGLEWMGW ISTYNGNTNY AQKFQGRVTM TTDTSTTTGY MELRRLRSD DTAVYYCARD YTRGAWFGES LIGGFDNWGQ GTLVTVSSAS TKGPSVFPLA PSSKSTSGGT AALGCLVKDY F PEPVTVSW ...String: QVQLVQSGAE VKKPGASVKV SCKASGYPFT SYGISWVRQA PGQGLEWMGW ISTYNGNTNY AQKFQGRVTM TTDTSTTTGY MELRRLRSD DTAVYYCARD YTRGAWFGES LIGGFDNWGQ GTLVTVSSAS TKGPSVFPLA PSSKSTSGGT AALGCLVKDY F PEPVTVSW NSGALTSGVH TFPAVLQSSG LYSLSSVVTV PSSSLGTQTY ICNVNHKPSN TKVDKKVEPK SC |
-Macromolecule #2: S309 light chain
| Macromolecule | Name: S309 light chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.204697 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPGT LSLSPGERAT LSCRASQTVS STSLAWYQQK PGQAPRLLIY GASSRATGIP DRFSGSGSGT DFTLTISRLE PEDFAVYYC QQHDTSLTFG GGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...String: EIVLTQSPGT LSLSPGERAT LSCRASQTVS STSLAWYQQK PGQAPRLLIY GASSRATGIP DRFSGSGSGT DFTLTISRLE PEDFAVYYC QQHDTSLTFG GGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC |
-Macromolecule #3: Spike protein S1
| Macromolecule | Name: Spike protein S1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30.444469 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: PTESIVRFPN ITNLCPFDEV FNATRFASVY AWNRKRISNC VADYSVLYNL APFFTFKCYG VSPTKLNDLC FTNVYADSFV IRGDEVRQI APGQTGNIAD YNYKLPDDFT GCVIAWNSNK LDSKVSGNYN YLYRLFRKSN LKPFERDIST EIYQAGNKPC N GVAGFNCY ...String: PTESIVRFPN ITNLCPFDEV FNATRFASVY AWNRKRISNC VADYSVLYNL APFFTFKCYG VSPTKLNDLC FTNVYADSFV IRGDEVRQI APGQTGNIAD YNYKLPDDFT GCVIAWNSNK LDSKVSGNYN YLYRLFRKSN LKPFERDIST EIYQAGNKPC N GVAGFNCY FPLRSYSFRP TYGVGHQPYR VVVLSFELLH APATVCGPKK STNLVKNKCV NFNFNGLKGT GVLTESNKKF LP FQQFGRD IADTTDAVRD PQTLEILDIT PC UniProtKB: Spike glycoprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.39 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)