[English] 日本語
 Yorodumi
Yorodumi- EMDB-33124: Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR)... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with S309 fab | |||||||||
 Map data Map data | Cryo-EM structure of SARS-CoV-2 Omicron spike protein in complex with S309 fab | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Omicron / spike / SARS-CoV-2 / S309 / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Gao GF / Qi JX / Zhao ZN / Liu S / Xie YF | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape Authors: Zhao Z / Zhou J / Tian M / Huang M / Liu S / Xie Y / Han P / Bai C / Han P / Zheng A / Fu L / Gao Y / Peng Q / Li Y / Chai Y / Zhang Z / Zhao X / Song H / Qi J / Wang Q / Wang P / Gao GF | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33124.map.gz emd_33124.map.gz | 483 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33124-v30.xml emd-33124-v30.xml emd-33124.xml emd-33124.xml | 21.2 KB 21.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33124.png emd_33124.png | 99.7 KB | ||
| Filedesc metadata |  emd-33124.cif.gz emd-33124.cif.gz | 7 KB | ||
| Others |  emd_33124_half_map_1.map.gz emd_33124_half_map_1.map.gz emd_33124_half_map_2.map.gz emd_33124_half_map_2.map.gz | 474.5 MB 474.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33124 http://ftp.pdbj.org/pub/emdb/structures/EMD-33124 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33124 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33124 | HTTPS FTP |
-Related structure data
| Related structure data |  7xcoMC  7xchC  7xciC  7xckC  7xcpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33124.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33124.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SARS-CoV-2 Omicron spike protein in complex with S309 fab | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.67 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Cryo-EM structure of SARS-CoV-2 Omicron spike protein in...
| File | emd_33124_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SARS-CoV-2 Omicron spike protein in complex with S309 fab | ||||||||||||
| Projections & Slices |
| ||||||||||||
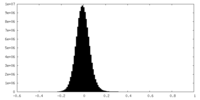
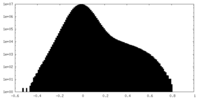
| Density Histograms |
-Half map: Cryo-EM structure of SARS-CoV-2 Omicron spike protein in...
| File | emd_33124_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SARS-CoV-2 Omicron spike protein in complex with S309 fab | ||||||||||||
| Projections & Slices |
| ||||||||||||
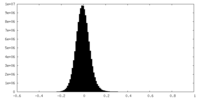
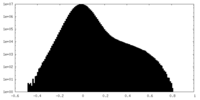
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of SARS-CoV-2 Omicron spike protein in complex ...
| Entire | Name: Cryo-EM structure of SARS-CoV-2 Omicron spike protein in complex with S309 fab |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of SARS-CoV-2 Omicron spike protein in complex ...
| Supramolecule | Name: Cryo-EM structure of SARS-CoV-2 Omicron spike protein in complex with S309 fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: SARS-CoV-2 Omicron spike protein
| Supramolecule | Name: SARS-CoV-2 Omicron spike protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: S309 fab
| Supramolecule | Name: S309 fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 138.306484 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPFFSNVT WFHVISGTNG TKRFDNPVLP FNDGVYFASI EKSNIIRGW IFGTTLDSKT QSLLIVNNAT NVVIKVCEFQ FCNDPFLDHK NNKSWMESEF RVYSSANNCT FEYVSQPFLM D LEGKQGNF ...String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPFFSNVT WFHVISGTNG TKRFDNPVLP FNDGVYFASI EKSNIIRGW IFGTTLDSKT QSLLIVNNAT NVVIKVCEFQ FCNDPFLDHK NNKSWMESEF RVYSSANNCT FEYVSQPFLM D LEGKQGNF KNLREFVFKN IDGYFKIYSK HTPIIVREPE DLPQGFSALE PLVDLPIGIN ITRFQTLLAL HRSYLTPGDS SS GWTAGAA AYYVGYLQPR TFLLKYNENG TITDAVDCAL DPLSETKCTL KSFTVEKGIY QTSNFRVQPT ESIVRFPNIT NLC PFDEVF NATRFASVYA WNRKRISNCV ADYSVLYNLA PFFTFKCYGV SPTKLNDLCF TNVYADSFVI RGDEVRQIAP GQTG NIADY NYKLPDDFTG CVIAWNSNKL DSKVSGNYNY LYRLFRKSNL KPFERDISTE IYQAGNKPCN GVAGFNCYFP LRSYS FRPT YGVGHQPYRV VVLSFELLHA PATVCGPKKS TNLVKNKCVN FNFNGLKGTG VLTESNKKFL PFQQFGRDIA DTTDAV RDP QTLEILDITP CSFGGVSVIT PGTNTSNQVA VLYQGVNCTE VPVAIHADQL TPTWRVYSTG SNVFQTRAGC LIGAEYV NN SYECDIPIGA GICASYQTQT KSHRRARSVA SQSIIAYTMS LGAENSVAYS NNSIAIPTNF TISVTTEILP VSMTKTSV D CTMYICGDST ECSNLLLQYG SFCTQLKRAL TGIAVEQDKN TQEVFAQVKQ IYKTPPIKYF GGFNFSQILP DPSKPSKRS PIEDLLFNKV TLADAGFIKQ YGDCLGDIAA RDLICAQKFK GLTVLPPLLT DEMIAQYTSA LLAGTITSGW TFGAGPALQI PFPMQMAYR FNGIGVTQNV LYENQKLIAN QFNSAIGKIQ DSLSSTPSAL GKLQDVVNHN AQALNTLVKQ LSSKFGAISS V LNDIFSRL DPPEAEVQID RLITGRLQSL QTYVTQQLIR AAEIRASANL AATKMSECVL GQSKRVDFCG KGYHLMSFPQ SA PHGVVFL HVTYVPAQEK NFTTAPAICH DGKAHFPREG VFVSNGTHWF VTQRNFYEPQ IITTDNTFVS GNCDVVIGIV NNT VYDPLQ PELDSFKEEL DKYFKNHTSP DVDLGDISGI NASVVNIQKE IDRLNEVAKN LNESLIDLQE LGKYEQGSGY IPEA PRDGQ AYVRKDGEWV LLSTFLGSAW SHPQFEKHHH HHHHH UniProtKB: Spike glycoprotein |
-Macromolecule #2: S309 Fab heavy chain
| Macromolecule | Name: S309 Fab heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.573471 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE VKKPGASVKV SCKASGYPFT SYGISWVRQA PGQGLEWMGW ISTYNGNTNY AQKFQGRVTM TTDTSTTTGY MELRRLRSD DTAVYYCARD YTRGAWFGES LIGGFDNWGQ GTLVTVSSAS TKGPSVFPLA PSSKSTSGGT AALGCLVKDY F PEPVTVSW ...String: QVQLVQSGAE VKKPGASVKV SCKASGYPFT SYGISWVRQA PGQGLEWMGW ISTYNGNTNY AQKFQGRVTM TTDTSTTTGY MELRRLRSD DTAVYYCARD YTRGAWFGES LIGGFDNWGQ GTLVTVSSAS TKGPSVFPLA PSSKSTSGGT AALGCLVKDY F PEPVTVSW NSGALTSGVH TFPAVLQSSG LYSLSSVVTV PSSSLGTQTY ICNVNHKPSN TKVDKKVEPK SC |
-Macromolecule #3: S309 fab light chain
| Macromolecule | Name: S309 fab light chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.204697 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPGT LSLSPGERAT LSCRASQTVS STSLAWYQQK PGQAPRLLIY GASSRATGIP DRFSGSGSGT DFTLTISRLE PEDFAVYYC QQHDTSLTFG GGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...String: EIVLTQSPGT LSLSPGERAT LSCRASQTVS STSLAWYQQK PGQAPRLLIY GASSRATGIP DRFSGSGSGT DFTLTISRLE PEDFAVYYC QQHDTSLTFG GGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 25 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.39 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)