+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Protein 110 and 12 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationenergy reserve metabolic process / fat cell differentiation / regulation of lipid metabolic process / synapse assembly / G protein-coupled receptor activity / memory / Olfactory Signaling Pathway / neuron projection development / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade ...energy reserve metabolic process / fat cell differentiation / regulation of lipid metabolic process / synapse assembly / G protein-coupled receptor activity / memory / Olfactory Signaling Pathway / neuron projection development / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / GTPase binding / cytoplasmic vesicle / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Ras protein signal transduction / cell surface receptor signaling pathway / Extra-nuclear estrogen signaling / cell population proliferation / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / synapse / protein-containing complex binding / signal transduction / extracellular exosome / extracellular region / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
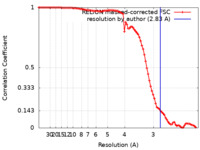
| Method | single particle reconstruction / cryo EM / Resolution: 2.83 Å | |||||||||
 Authors Authors | He Y / Zhu X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural basis of adhesion GPCR GPR110 activation by stalk peptide and G-proteins coupling. Authors: Xinyan Zhu / Yu Qian / Xiaowan Li / Zhenmei Xu / Ruixue Xia / Na Wang / Jiale Liang / Han Yin / Anqi Zhang / Changyou Guo / Guangfu Wang / Yuanzheng He /  Abstract: Adhesion G protein-coupled receptors (aGPCRs) are keys of many physiological events and attractive targets for various diseases. aGPCRs are also known to be capable of self-activation via an ...Adhesion G protein-coupled receptors (aGPCRs) are keys of many physiological events and attractive targets for various diseases. aGPCRs are also known to be capable of self-activation via an autoproteolysis process that removes the inhibitory GAIN domain on the extracellular side of receptor and releases a stalk peptide to bind and activate the transmembrane side of receptor. However, the detailed mechanism of aGPCR activation remains elusive. Here, we report the cryo-electron microscopy structures of GPR110 (ADGRF1), a member of aGPCR, in complex with G, G, G, G and G The structures reveal distinctive ligand engaging model and activation conformations of GPR110. The structures also unveil the rarely explored GPCR/G and GPCR/G engagements. A comparison of G, G, G, G and G engagements with GPR110 reveals details of G-protein engagement, including a dividing point at the far end of the alpha helix 5 (αH5) of Gα subunit that separates G/G engagements from G/G/G engagements. This is also where G/G bind the receptor through both hydrophobic and polar interaction, while G/G/G engage receptor mainly through hydrophobic interaction. We further provide physiological evidence of GPR110 activation via stalk peptide. Taken together, our study fills the missing information of GPCR/G-protein engagement and provides a framework for understanding aGPCR activation and GPR110 signaling. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32905.map.gz emd_32905.map.gz | 55.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32905-v30.xml emd-32905-v30.xml emd-32905.xml emd-32905.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32905_fsc.xml emd_32905_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_32905.png emd_32905.png | 70.3 KB | ||
| Filedesc metadata |  emd-32905.cif.gz emd-32905.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32905 http://ftp.pdbj.org/pub/emdb/structures/EMD-32905 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32905 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32905 | HTTPS FTP |
-Related structure data
| Related structure data |  7wz7MC  7wxuC  7wxwC  7wy0C  7x2vC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32905.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32905.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : GPCR/G-protein complex
| Entire | Name: GPCR/G-protein complex |
|---|---|
| Components |
|
-Supramolecule #1: GPCR/G-protein complex
| Supramolecule | Name: GPCR/G-protein complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: engineered G alpha 12 subunit
| Macromolecule | Name: engineered G alpha 12 subunit / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.957145 KDa |
| Recombinant expression | Organism:  Insect BA phytoplasma (bacteria) Insect BA phytoplasma (bacteria) |
| Sequence | String: MMGCTLSAED KAAVERSKMI DALLARERRA VRRLVKILLL GADNSGKSTF LKQMRIIHVN GYSEEECKQY KAVVYSNTIQ SIIAIIRAM GRLKIDFGDS ARADDARQLF VLAGAAEEGF MTAELAGVIK RLWKDSGVQA CFNRSREYQL NDSAAYYLND L DRIAQPNY ...String: MMGCTLSAED KAAVERSKMI DALLARERRA VRRLVKILLL GADNSGKSTF LKQMRIIHVN GYSEEECKQY KAVVYSNTIQ SIIAIIRAM GRLKIDFGDS ARADDARQLF VLAGAAEEGF MTAELAGVIK RLWKDSGVQA CFNRSREYQL NDSAAYYLND L DRIAQPNY IPTQQDVLRT RVKTKGIVEH DFVIKKIPFK MVDVGAQRSQ RQKWFQCFDG ITSILFMVDS SDYNRLVESM ND FETIVNN KLFFNVSIIL FLNKMDLLVE KVKTVSIKKH FPDFRGDPHR LEDVQRYLVQ CFDRKRRNRS KPLFHHFTTS IDT ENARFI FHAVKDTILQ ENLKDIMLQ |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.915496 KDa |
| Recombinant expression | Organism:  Insect BA phytoplasma (bacteria) Insect BA phytoplasma (bacteria) |
| Sequence | String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD ...String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD TTCALWDIET GQQTTTFTGH TGDVMSLSLA PDTRLFVSGA CDASAKLWDV REGMCRQTFT GHESDINAIC FF PNGNAFA TGSDDATCRL FDLRADQELM TYSHDNIICG ITSVSFSKSG RLLLAGYDDF NCNVWDALKA DRAGVLAGHD NRV SCLGVT DDGMAVATGS WDSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.277299 KDa |
| Recombinant expression | Organism:  Insect BA phytoplasma (bacteria) Insect BA phytoplasma (bacteria) |
| Sequence | String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSADIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSADIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  Insect BA phytoplasma (bacteria) Insect BA phytoplasma (bacteria) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: Adhesion G-protein coupled receptor F1
| Macromolecule | Name: Adhesion G-protein coupled receptor F1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 101.465219 KDa |
| Recombinant expression | Organism:  Insect BA phytoplasma (bacteria) Insect BA phytoplasma (bacteria) |
| Sequence | String: MKVGVLWLIS FFTFTDGHGG FLGKNDGIKT KKELIVNKKK HLGPVEEYQL LLQVTYRDSK EKRDLRNFLK LLKPPLLWSH GLIRIIRAK ATTDCNSLNG VLQCTCEDSY TWFPPSCLDP QNCYLHTAGA LPSCECHLNN LSQSVNFCER TKIWGTFKIN E RFTNDLLN ...String: MKVGVLWLIS FFTFTDGHGG FLGKNDGIKT KKELIVNKKK HLGPVEEYQL LLQVTYRDSK EKRDLRNFLK LLKPPLLWSH GLIRIIRAK ATTDCNSLNG VLQCTCEDSY TWFPPSCLDP QNCYLHTAGA LPSCECHLNN LSQSVNFCER TKIWGTFKIN E RFTNDLLN SSSAIYSKYA NGIEIQLKKA YERIQGFESV QVTQFRNGSI VAGYEVVGSS SASELLSAIE HVAEKAKTAL HK LFPLEDG SFRVFGKAQC NDIVFGFGSK DDEYTLPCSS GYRGNITAKC ESSGWQVIRE TCVLSLLEEL NKNFSMIVGN ATE AAVSSF VQNLSVIIRQ NPSTTVGNLA SVVSILSNIS SLSLASHFRV SNSTMEDVIS IADNILNSAS VTNWTVLLRE EKYA SSRLL ETLENISTLV PPTALPLNFS RKFIDWKGIP VNKSQLKRGY SYQIKMCPQN TSIPIRGRVL IGSDQFQRSL PETII SMAS LTLGNILPVS KNGNAQVNGP VISTVIQNYS INEVFLFFSK IESNLSQPHC VFWDFSHLQW NDAGCHLVNE TQDIVT CQC THLTSFSILM SPFVPSTIFP VVKWITYVGL GISIGSLILC LIIEALFWKQ IKKSQTSHTR RICMVNIALS LLIADVW FI VGATVDTTVN PSGVCTAAVF FTHFFYLSLF FWMLMLGILL AYRIILVFHH MAQHLMMAVG FCLGYGCPLI ISVITIAV T QPSNTYKRKD VCWLNWSNGS KPLLAFVVPA LAIVAVNFVV VLLVLTKLWR PTVGERLSRD DKATIIRVGK SLLILTPLL GLTWGFGIGT IVDSQNLAWH VIFALLNAFQ GFFILCFGIL LDSKLRQLLF NKLSALSSWK QTEKQNSSDL SAKPKFSKPF NPLQNKGHY AFSHTGDSSD NIMLTQFVSN E UniProtKB: Adhesion G-protein coupled receptor F1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)