[English] 日本語
 Yorodumi
Yorodumi- EMDB-32582: Cryo-EM structure of the human glycosylphosphatidylinositol trans... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human glycosylphosphatidylinositol transamidase complex at 2.53 Angstrom resolution | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationGPI-anchor transamidase activity / attachment of GPI anchor to protein / GPI-anchor transamidase complex / GPI anchor biosynthetic process / protein retention in ER lumen / Attachment of GPI anchor to uPAR / Hydrolases / regulation of receptor signaling pathway via JAK-STAT / protein disulfide isomerase activity / tubulin binding ...GPI-anchor transamidase activity / attachment of GPI anchor to protein / GPI-anchor transamidase complex / GPI anchor biosynthetic process / protein retention in ER lumen / Attachment of GPI anchor to uPAR / Hydrolases / regulation of receptor signaling pathway via JAK-STAT / protein disulfide isomerase activity / tubulin binding / neuron differentiation / cytoplasmic vesicle / protein-containing complex assembly / neuron apoptotic process / centrosome / endoplasmic reticulum membrane / endoplasmic reticulum / mitochondrion / proteolysis / membrane / plasma membrane / cytosol Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.53 Å | ||||||||||||||||||
 Authors Authors | Xu Y / Li T / Luo Y / Chao Y / Jia G / Zhou Z / Su Z / Qu Q / Li D | ||||||||||||||||||
| Funding support |  China, 5 items China, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Molecular insights into biogenesis of glycosylphosphatidylinositol anchor proteins. Authors: Yidan Xu / Guowen Jia / Tingting Li / Zixuan Zhou / Yitian Luo / Yulin Chao / Juan Bao / Zhaoming Su / Qianhui Qu / Dianfan Li /  Abstract: Eukaryotic cells are coated with an abundance of glycosylphosphatidylinositol anchor proteins (GPI-APs) that play crucial roles in fertilization, neurogenesis, and immunity. The removal of a ...Eukaryotic cells are coated with an abundance of glycosylphosphatidylinositol anchor proteins (GPI-APs) that play crucial roles in fertilization, neurogenesis, and immunity. The removal of a hydrophobic signal peptide and covalent attachment of GPI at the new carboxyl terminus are catalyzed by an endoplasmic reticulum membrane GPI transamidase complex (GPI-T) conserved among all eukaryotes. Here, we report the cryo-electron microscopy (cryo-EM) structure of the human GPI-T at a global 2.53-Å resolution, revealing an equimolar heteropentameric assembly. Structure-based mutagenesis suggests a legumain-like mechanism for the recognition and cleavage of proprotein substrates, and an endogenous GPI in the structure defines a composite cavity for the lipid substrate. This elongated active site, stemming from the membrane and spanning an additional ~22-Å space toward the catalytic dyad, is structurally suited for both substrates which feature an amphipathic pattern that matches this geometry. Our work presents an important step towards the mechanistic understanding of GPI-AP biosynthesis. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32582.map.gz emd_32582.map.gz | 79 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32582-v30.xml emd-32582-v30.xml emd-32582.xml emd-32582.xml | 28.5 KB 28.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32582.png emd_32582.png | 82.6 KB | ||
| Others |  emd_32582_additional_1.map.gz emd_32582_additional_1.map.gz | 42.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32582 http://ftp.pdbj.org/pub/emdb/structures/EMD-32582 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32582 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32582 | HTTPS FTP |
-Validation report
| Summary document |  emd_32582_validation.pdf.gz emd_32582_validation.pdf.gz | 505.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32582_full_validation.pdf.gz emd_32582_full_validation.pdf.gz | 504.7 KB | Display | |
| Data in XML |  emd_32582_validation.xml.gz emd_32582_validation.xml.gz | 6.4 KB | Display | |
| Data in CIF |  emd_32582_validation.cif.gz emd_32582_validation.cif.gz | 7.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32582 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32582 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32582 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32582 | HTTPS FTP |
-Related structure data
| Related structure data |  7wldMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32582.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32582.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
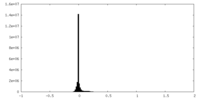
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: unshapened map
| File | emd_32582_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unshapened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
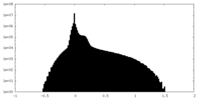
| Density Histograms |
- Sample components
Sample components
+Entire : GPI transamidase complex
+Supramolecule #1: GPI transamidase complex
+Macromolecule #1: Glycosylphosphatidylinositol anchor attachment 1 protein
+Macromolecule #2: GPI-anchor transamidase
+Macromolecule #3: GPI transamidase component PIG-S
+Macromolecule #4: GPI transamidase component PIG-T
+Macromolecule #5: Phosphatidylinositol glycan anchor biosynthesis class U protein
+Macromolecule #7: CHOLESTEROL HEMISUCCINATE
+Macromolecule #8: Digitonin
+Macromolecule #9: (4S,7R)-7-[(hexadecanoyloxy)methyl]-4-hydroxy-N,N,N-trimethyl-4,9...
+Macromolecule #10: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
+Macromolecule #11: CALCIUM ION
+Macromolecule #12: O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phospho...
+Macromolecule #13: 2-acetamido-2-deoxy-beta-D-glucopyranose
+Macromolecule #14: CHOLESTEROL
+Macromolecule #15: 2-azanylethyl [(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-...
+Macromolecule #16: 2-amino-2-deoxy-alpha-D-glucopyranose
+Macromolecule #17: CARDIOLIPIN
+Macromolecule #18: [(2R)-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy...
+Macromolecule #19: [(2~{R})-1-hexadecanoyloxy-3-[[(1~{S},2~{R},3~{R},4~{S},5~{S},6~{...
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 20 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.026000000000000002 kPa | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 53.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7wld: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)