+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | mouse Pendrin in bicarbonate and iodide buffer in inward state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | exchange / transport / slc / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationInorganic anion exchange by SLC26 transporters / iodide transmembrane transporter activity / secondary active sulfate transmembrane transporter activity / monoatomic anion transmembrane transporter activity / regulation of pH / chloride:bicarbonate antiporter activity / animal organ morphogenesis / brush border membrane / regulation of protein localization / apical plasma membrane ...Inorganic anion exchange by SLC26 transporters / iodide transmembrane transporter activity / secondary active sulfate transmembrane transporter activity / monoatomic anion transmembrane transporter activity / regulation of pH / chloride:bicarbonate antiporter activity / animal organ morphogenesis / brush border membrane / regulation of protein localization / apical plasma membrane / extracellular exosome / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.25 Å | |||||||||
 Authors Authors | Liu QY / Zhang X | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Asymmetric pendrin homodimer reveals its molecular mechanism as anion exchanger. Authors: Qianying Liu / Xiang Zhang / Hui Huang / Yuxin Chen / Fang Wang / Aihua Hao / Wuqiang Zhan / Qiyu Mao / Yuxia Hu / Lin Han / Yifang Sun / Meng Zhang / Zhimin Liu / Geng-Lin Li / Weijia Zhang ...Authors: Qianying Liu / Xiang Zhang / Hui Huang / Yuxin Chen / Fang Wang / Aihua Hao / Wuqiang Zhan / Qiyu Mao / Yuxia Hu / Lin Han / Yifang Sun / Meng Zhang / Zhimin Liu / Geng-Lin Li / Weijia Zhang / Yilai Shu / Lei Sun / Zhenguo Chen /  Abstract: Pendrin (SLC26A4) is an anion exchanger expressed in the apical membranes of selected epithelia. Pendrin ablation causes Pendred syndrome, a genetic disorder associated with sensorineural hearing ...Pendrin (SLC26A4) is an anion exchanger expressed in the apical membranes of selected epithelia. Pendrin ablation causes Pendred syndrome, a genetic disorder associated with sensorineural hearing loss, hypothyroid goiter, and reduced blood pressure. However its molecular structure has remained unknown, limiting our understanding of the structural basis of transport. Here, we determine the cryo-electron microscopy structures of mouse pendrin with symmetric and asymmetric homodimer conformations. The asymmetric homodimer consists of one inward-facing protomer and the other outward-facing protomer, representing coincident uptake and secretion- a unique state of pendrin as an electroneutral exchanger. The multiple conformations presented here provide an inverted alternate-access mechanism for anion exchange. The structural and functional data presented here disclose the properties of an anion exchange cleft and help understand the importance of disease-associated variants, which will shed light on the pendrin exchange mechanism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32574.map.gz emd_32574.map.gz | 110.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32574-v30.xml emd-32574-v30.xml emd-32574.xml emd-32574.xml | 12.8 KB 12.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32574.png emd_32574.png | 116.5 KB | ||
| Filedesc metadata |  emd-32574.cif.gz emd-32574.cif.gz | 6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32574 http://ftp.pdbj.org/pub/emdb/structures/EMD-32574 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32574 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32574 | HTTPS FTP |
-Related structure data
| Related structure data |  7wl2MC  7wk1C  7wk7C  7wl7C  7wl8C  7wl9C  7wlaC  7wlbC  7wleC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32574.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32574.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.046 Å | ||||||||||||||||||||||||||||||||||||
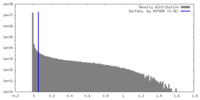
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : mouse Pendrin in bicarbonate and iodide buffer in inward state
| Entire | Name: mouse Pendrin in bicarbonate and iodide buffer in inward state |
|---|---|
| Components |
|
-Supramolecule #1: mouse Pendrin in bicarbonate and iodide buffer in inward state
| Supramolecule | Name: mouse Pendrin in bicarbonate and iodide buffer in inward state type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Pendrin
| Macromolecule | Name: Pendrin / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 85.769578 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAARGGRSEP PQLAEYSCSY TVSRPVYSEL AFQQQRERRL PERRTLRDSL ARSCSCSRKR AFGVVKTLLP ILDWLPKYRV KEWLLSDII SGVSTGLVGT LQGMAYALLA AVPVQFGLYS AFFPILTYFV FGTSRHISVG PFPVVSLMVG SVVLSMAPDD H FLVPSGNG ...String: MAARGGRSEP PQLAEYSCSY TVSRPVYSEL AFQQQRERRL PERRTLRDSL ARSCSCSRKR AFGVVKTLLP ILDWLPKYRV KEWLLSDII SGVSTGLVGT LQGMAYALLA AVPVQFGLYS AFFPILTYFV FGTSRHISVG PFPVVSLMVG SVVLSMAPDD H FLVPSGNG SALNSTTLDT GTRDAARVLL ASTLTLLVGI IQLVFGGLQI GFIVRYLADP LVGGFTTAAA FQVLVSQLKI VL NVSTKNY NGILSIIYTL IEIFQNIGDT NIADFIAGLL TIIVCMAVKE LNDRFKHRIP VPIPIEVIVT IIATAISYGA NLE KNYNAG IVKSIPSGFL PPVLPSVGLF SDMLAASFSI AVVAYAIAVS VGKVYATKHD YVIDGNQEFI AFGISNVFSG FFSC FVATT ALSRTAVQES TGGKTQVAGL ISAVIVMVAI VALGRLLEPL QKSVLAAVVI ANLKGMFMQV CDVPRLWKQN KTDAV IWVF TCIMSIILGL DLGLLAGLLF ALLTVVLRVQ FPSWNGLGSV PSTDIYKSIT HYKNLEEPEG VKILRFSSPI FYGNVD GFK KCINSTVGFD AIRVYNKRLK ALRRIQKLIK KGQLRATKNG IISDIGSSNN AFEPDEDVEE PEELNIPTKE IEIQVDW NS ELPVKVNVPK VPIHSLVLDC GAVSFLDVVG VRSLRMIVKE FQRIDVNVYF ALLQDDVLEK MEQCGFFDDN IRKDRFFL T VHDAILHLQN QVKSREGQDS LLETVARIRD CKDPLDLMEA EMNAEELDVQ DEAMRRLAS UniProtKB: Pendrin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 53.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)