[English] 日本語
 Yorodumi
Yorodumi- EMDB-31676: Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-b... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31676 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GCGR-Gs complex | ||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of glycogen metabolic process / glucagon receptor activity / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding ...regulation of glycogen metabolic process / glucagon receptor activity / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Ca2+ pathway / G alpha (z) signalling events / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / G alpha (q) signalling events / Thrombin signalling through proteinase activated receptors (PARs) / G alpha (i) signalling events / alkylglycerophosphoethanolamine phosphodiesterase activity / Activation of G protein gated Potassium channels / G-protein activation / G beta:gamma signalling through PI3Kgamma / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through PLC beta / ADP signalling through P2Y purinoceptor 1 / Thromboxane signalling through TP receptor / Presynaptic function of Kainate receptors / G beta:gamma signalling through CDC42 / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Glucagon-type ligand receptors / G alpha (12/13) signalling events / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Adrenaline,noradrenaline inhibits insulin secretion / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Thrombin signalling through proteinase activated receptors (PARs) / Ca2+ pathway / Extra-nuclear estrogen signaling / G alpha (z) signalling events / G alpha (s) signalling events / G alpha (q) signalling events / photoreceptor outer segment membrane / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (i) signalling events / spectrin binding / Vasopressin regulates renal water homeostasis via Aquaporins / response to starvation / exocytosis / PKA activation in glucagon signalling / peptide hormone binding / hair follicle placode formation / developmental growth / photoreceptor outer segment / D1 dopamine receptor binding / intracellular transport / renal water homeostasis / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway / activation of adenylate cyclase activity / cardiac muscle cell apoptotic process / cellular response to glucagon stimulus / adenylate cyclase activator activity / hormone-mediated signaling pathway / cellular response to starvation / regulation of insulin secretion / photoreceptor inner segment / response to nutrient / trans-Golgi network membrane / guanyl-nucleotide exchange factor activity / generation of precursor metabolites and energy / negative regulation of inflammatory response to antigenic stimulus / bone development / G-protein beta/gamma-subunit complex binding / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / G protein activity / platelet aggregation / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / regulation of blood pressure / cognition / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / sensory perception of taste / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / cellular response to prostaglandin E stimulus / G-protein beta-subunit binding / heterotrimeric G-protein complex / sensory perception of smell / signaling receptor complex adaptor activity / GTPase binding / positive regulation of cold-induced thermogenesis Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   | ||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | ||||||||||||||||||||||||
 Authors Authors | Zhao F / Zhou QT / Cong ZT / Hang KN / Zou XY / Zhang C / Chen Y / Dai AT / Liang AY / Ming QQ ...Zhao F / Zhou QT / Cong ZT / Hang KN / Zou XY / Zhang C / Chen Y / Dai AT / Liang AY / Ming QQ / Wang M / Chen LN / Xu PY / Chang RL / Feng WB / Xia T / Zhang Y / Wu BL / Yang DH / Zhao LH / Xu HE / Wang MW | ||||||||||||||||||||||||
| Funding support |  China, 7 items China, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors. Authors: Fenghui Zhao / Qingtong Zhou / Zhaotong Cong / Kaini Hang / Xinyu Zou / Chao Zhang / Yan Chen / Antao Dai / Anyi Liang / Qianqian Ming / Mu Wang / Li-Nan Chen / Peiyu Xu / Rulve Chang / ...Authors: Fenghui Zhao / Qingtong Zhou / Zhaotong Cong / Kaini Hang / Xinyu Zou / Chao Zhang / Yan Chen / Antao Dai / Anyi Liang / Qianqian Ming / Mu Wang / Li-Nan Chen / Peiyu Xu / Rulve Chang / Wenbo Feng / Tian Xia / Yan Zhang / Beili Wu / Dehua Yang / Lihua Zhao / H Eric Xu / Ming-Wei Wang /  Abstract: Glucose homeostasis, regulated by glucose-dependent insulinotropic polypeptide (GIP), glucagon-like peptide-1 (GLP-1) and glucagon (GCG) is critical to human health. Several multi-targeting agonists ...Glucose homeostasis, regulated by glucose-dependent insulinotropic polypeptide (GIP), glucagon-like peptide-1 (GLP-1) and glucagon (GCG) is critical to human health. Several multi-targeting agonists at GIPR, GLP-1R or GCGR, developed to maximize metabolic benefits with reduced side-effects, are in clinical trials to treat type 2 diabetes and obesity. To elucidate the molecular mechanisms by which tirzepatide, a GIPR/GLP-1R dual agonist, and peptide 20, a GIPR/GLP-1R/GCGR triagonist, manifest their multiplexed pharmacological actions over monoagonists such as semaglutide, we determine cryo-electron microscopy structures of tirzepatide-bound GIPR and GLP-1R as well as peptide 20-bound GIPR, GLP-1R and GCGR. The structures reveal both common and unique features for the dual and triple agonism by illustrating key interactions of clinical relevance at the near-atomic level. Retention of glucagon function is required to achieve such an advantage over GLP-1 monotherapy. Our findings provide valuable insights into the structural basis of functional versatility of tirzepatide and peptide 20. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31676.map.gz emd_31676.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31676-v30.xml emd-31676-v30.xml emd-31676.xml emd-31676.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31676.png emd_31676.png | 18.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31676 http://ftp.pdbj.org/pub/emdb/structures/EMD-31676 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31676 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31676 | HTTPS FTP |
-Validation report
| Summary document |  emd_31676_validation.pdf.gz emd_31676_validation.pdf.gz | 404.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31676_full_validation.pdf.gz emd_31676_full_validation.pdf.gz | 403.7 KB | Display | |
| Data in XML |  emd_31676_validation.xml.gz emd_31676_validation.xml.gz | 6.2 KB | Display | |
| Data in CIF |  emd_31676_validation.cif.gz emd_31676_validation.cif.gz | 7.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31676 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31676 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31676 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31676 | HTTPS FTP |
-Related structure data
| Related structure data |  7v35MC  7fimC  7finC  7fiyC  7vabC  7vbhC  7vbiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31676.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31676.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.071 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
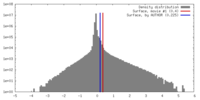
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-b...
| Entire | Name: Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GCGR-Gs complex |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-b...
| Supramolecule | Name: Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GCGR-Gs complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
-Macromolecule #1: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.683434 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKNTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE ...String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKNTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE YQLIDCAQYF LDKIDVIKQA DYVPSDQDLL RCRVLTSGIF ETKFQVDKVN FHMFDVGAQR DERRKWIQCF ND VTAIIFV VASSSYNMVI REDNQTNRLQ AALKLFDSIW NNKWLRDTSV ILFLNKQDLL AEKVLAGKSK IEDYFPEFAR YTT PEDATP EPGEDPRVTR AKYFIRDEFL RISTASGDGR HYCYPHFTCA VDTENIRRVF NDCRDIIQRM HLRQYELL |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.915496 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD ...String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD TTCALWDIET GQQTTTFTGH TGDVMSLSLA PDTRLFVSGA CDASAKLWDV REGMCRQTFT GHESDINAIC FF PNGNAFA TGSDDATCRL FDLRADQELM TYSHDNIICG ITSVSFSKSG RLLLAGYDDF NCNVWDALKA DRAGVLAGHD NRV SCLGVT DDGMAVATGS WDSFLKIWN |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L |
-Macromolecule #4: Nanobody-35
| Macromolecule | Name: Nanobody-35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 15.343019 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAQVQLQESG GGLVQPGGSL RLSCAASGFT FSNYKMNWVR QAPGKGLEWV SDISQSGASI SYTGSVKGRF TISRDNAKNT LYLQMNSLK PEDTAVYYCA RCPAPFTRDC FDVTSTTYAY RGQGTQVTVS SHHHHHHEPE A |
-Macromolecule #5: Peptide 20
| Macromolecule | Name: Peptide 20 / type: protein_or_peptide / ID: 5 Details: At positon of C10 attached PL2=K(gE-C16), lysine with a gamaE-C16 acyl which is attached through the side chain amine. And there is a NH2 at the C terminus of the peptide. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 4.180502 KDa |
| Sequence | String: H(AIB)QGTFTSDK SKYLDERAAQ DFVQWLLDGG PSSGAPPPS |
-Macromolecule #6: Glucagon receptor
| Macromolecule | Name: Glucagon receptor / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 46.741363 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVMDFLFEKW KLYGDQCHHN LSLLPPPTEL VCNRTFDKYS CWPDTPANTT ANISCPWYLP WHHKVQHRFV FKRCGPDGQW VRGPRGQPW RDASQCQMDG EEIEVQKEVA KMYSSFQVMY TVGYSLSLGA LLLALAILGG LSKLHCTRNA IHANLFASFV L KASSVLVI ...String: QVMDFLFEKW KLYGDQCHHN LSLLPPPTEL VCNRTFDKYS CWPDTPANTT ANISCPWYLP WHHKVQHRFV FKRCGPDGQW VRGPRGQPW RDASQCQMDG EEIEVQKEVA KMYSSFQVMY TVGYSLSLGA LLLALAILGG LSKLHCTRNA IHANLFASFV L KASSVLVI DGLLRTRYSQ KIGDDLSVST WLSDGAVAGC RVAAVFMQYG IVANYCWLLV EGLYLHNLLG LATLPERSFF SL YLGIGWG APMLFVVPWA VVKCLFENVQ CWTSNDNMGF WWILRFPVFL AILINFFIFV RIVQLLVAKL RARQMHHTDY KFR LAKSTL TLIPLLGVHE VVFAFVTDEH AQGTLRSAKL FFDLFLSSFQ GLLVAVLYCF LNKEVQSELR RRWHRWRLGK VLWE ERNTS N |
-Macromolecule #7: N-hexadecanoyl-L-glutamic acid
| Macromolecule | Name: N-hexadecanoyl-L-glutamic acid / type: ligand / ID: 7 / Number of copies: 1 / Formula: D6M |
|---|---|
| Molecular weight | Theoretical: 385.538 Da |
| Chemical component information |  ChemComp-D6M: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source: OTHER |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.5 Å / Resolution method: DIFFRACTION PATTERN/LAYERLINES / Number images used: 383657 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)