[English] 日本語
 Yorodumi
Yorodumi- EMDB-31199: Pyochelin synthetase, a dimeric nonribosomal peptide synthetase e... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31199 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Pyochelin synthetase, a dimeric nonribosomal peptide synthetase elongation module-after-condensation | |||||||||
 Map data Map data | PchE-after-condensation | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nonribosomal peptide synthetase / BIOSYNTHETIC PROTEIN / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationL-cysteine-[L-cysteinyl-carrier protein] ligase / ligase activity / phosphopantetheine binding / antibiotic biosynthetic process Similarity search - Function | |||||||||
| Biological species |  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) | |||||||||
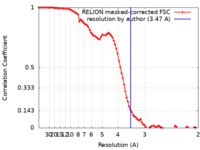
| Method | single particle reconstruction / cryo EM / Resolution: 3.47 Å | |||||||||
 Authors Authors | Wang JL / Wang ZJ | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Catalytic trajectory of a dimeric nonribosomal peptide synthetase subunit with an inserted epimerase domain. Authors: Jialiang Wang / Dandan Li / Lu Chen / Wei Cao / Liangliang Kong / Wei Zhang / Tristan Croll / Zixin Deng / Jingdan Liang / Zhijun Wang /   Abstract: Nonribosomal peptide synthetases (NRPSs) are modular assembly-line megaenzymes that synthesize diverse metabolites with wide-ranging biological activities. The structural dynamics of synthetic ...Nonribosomal peptide synthetases (NRPSs) are modular assembly-line megaenzymes that synthesize diverse metabolites with wide-ranging biological activities. The structural dynamics of synthetic elongation has remained unclear. Here, we present cryo-EM structures of PchE, an NRPS elongation module, in distinct conformations. The domain organization reveals a unique "H"-shaped head-to-tail dimeric architecture. The capture of both aryl and peptidyl carrier protein-tethered substrates and intermediates inside the heterocyclization domain and L-cysteinyl adenylate in the adenylation domain illustrates the catalytic and recognition residues. The multilevel structural transitions guided by the adenylation C-terminal subdomain in combination with the inserted epimerase and the conformational changes of the heterocyclization tunnel are controlled by two residues. Moreover, we visualized the direct structural dynamics of the full catalytic cycle from thiolation to epimerization. This study establishes the catalytic trajectory of PchE and sheds light on the rational re-engineering of domain-inserted dimeric NRPSs for the production of novel pharmaceutical agents. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31199.map.gz emd_31199.map.gz | 46.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31199-v30.xml emd-31199-v30.xml emd-31199.xml emd-31199.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31199_fsc.xml emd_31199_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_31199.png emd_31199.png | 68.4 KB | ||
| Others |  emd_31199_additional_1.map.gz emd_31199_additional_1.map.gz emd_31199_half_map_1.map.gz emd_31199_half_map_1.map.gz emd_31199_half_map_2.map.gz emd_31199_half_map_2.map.gz | 55.6 MB 46.2 MB 46.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31199 http://ftp.pdbj.org/pub/emdb/structures/EMD-31199 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31199 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31199 | HTTPS FTP |
-Related structure data
| Related structure data |  7en1MC  7emyC  7en2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31199.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31199.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PchE-after-condensation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
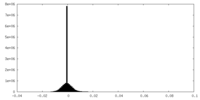
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: postprocess
| File | emd_31199_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | postprocess | ||||||||||||
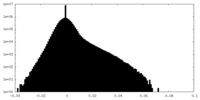
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half1
| File | emd_31199_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half2
| File | emd_31199_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : pyochelin synthetase, a dimeric nonribosomal peptide synthetase e...
| Entire | Name: pyochelin synthetase, a dimeric nonribosomal peptide synthetase elongation module |
|---|---|
| Components |
|
-Supramolecule #1: pyochelin synthetase, a dimeric nonribosomal peptide synthetase e...
| Supramolecule | Name: pyochelin synthetase, a dimeric nonribosomal peptide synthetase elongation module type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) |
| Molecular weight | Theoretical: 160 kDa/nm |
-Macromolecule #1: Dihydroaeruginoic acid synthetase
| Macromolecule | Name: Dihydroaeruginoic acid synthetase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria)Strain: ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1 |
| Molecular weight | Theoretical: 158.762641 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDLPPDSRTA LRDWLTEQLA DLLGEPLADV RALADDDDLL GCGLDSIRLM YLQERLRARG STLDFAQLAQ RPCLGAWLDL LACADRLSA PATVALPTAQ DRDQPFELSS VQQAYWLGRG AGEVLGNVSC HAFLEFRTRD VDPQRLAAAA ECVRQRHPML R ARFLDGRQ ...String: MDLPPDSRTA LRDWLTEQLA DLLGEPLADV RALADDDDLL GCGLDSIRLM YLQERLRARG STLDFAQLAQ RPCLGAWLDL LACADRLSA PATVALPTAQ DRDQPFELSS VQQAYWLGRG AGEVLGNVSC HAFLEFRTRD VDPQRLAAAA ECVRQRHPML R ARFLDGRQ QILPTPPLSC FDLQDWRTLQ VDEAERDWQA LRDWRAHECL AVERGQVFLL GLVRMPGGED RLWLSLDLLA AD VESLRLL LAELGVAYLA PERLAEPPAL HFADYLAHRA AQRAEAAARA RDYWLERLPR LPDAPALPLA CAPESIRQPR TRR LAFQLS AGESRRLERL AAQHGVTLSS VFGCAFALVL ARWSESAEFL LNVPLFDRHA DDPRIGEVIA DFTTLLLLEC RMQA GVSFA EAVKSFQRNL HGAIDHAAFP ALEVLREARR QGQPRSAPVV FASNLGEEGF VPAAFRDAFG DLHDMLSQTP QVWLD HQLY RVGDGILLAW DSVVGLFPEG LPETMFEAYV GLLQRLCDSA WGQPADLPLP WAQQARRALL NGQPACATAR TLHRDF FLR AAEAPDADAL LYRDQRVTRG ELAERALRIA GGLREAGVRP GDAVEVSLPR GPQQVAAVFG VLAAGACYVP LDIDQPP AR RRLIEEAAGV CLAITEEDDP QALPPRLDVQ RLLRGPALAA PVPLAPQASA YVIYTSGSTG VPKGVEVSHA AAINTIDA L LDLLRVNASD RLLAVSALDF DLSVFDLFGG LGAGASLVLP AQEQARDAAA WAEAIQRHAV SLWNSAPALL EMALSLPAS QADYRSLRAV LLSGDWVALD LPGRLRPRCA EGCRLHVLGG ATEAGIWSNL QSVDTVPPHW RSIPYGRPLP GQAYRVVDTH GRDVPDLVV GELWIGGASL ARGYRNDPEL SARRFVHDAQ GRWYRTGDRG RYWGDGTLEF LGRVDQQVKV RGQRIELGEV E AALCAQAG VESACAAVLG GGVASLGAVL VPRLAPRAEG SMDLPAAQPF AGLAEAEAVL TREILGALLE APLELDDGLR RR WLDWLAD SAASALPSLD EALRRLGWQA AGLTAMGNAL RGLLAGEQAP AALLLDPWLA PQAVAARLPD GREALARLLE ALP TPAAGE RLRVAVLDTR AGLWLDQGMA SLLRPGLELT LFERSRVLLD AAATRLPERI VVQALDDGLL PAEHLGRYDR VISF AALHA YEASREGLAL AAALLRPQGR LLLVDLLCES PLALLGAALL DDRPLRLAEL PSLLADLAAA GLAPRCLWRS ERIAL VEAL APGLGLDAAA LQAGLEQRLP QAMRPERLWC LPSLPLNGNG KVDRRRLAES MTRALGECRH EPSAEEPLEA HEQALA ECW EAVLKRPVRR REASFFSLGG DSLLATRLLA GIRERFGVRL GMADFYRQPT LAGLARHLQV QTVEIEETQL EEGVLHH HH HHLPSWSHPQ FEK |
-Macromolecule #2: 4'-PHOSPHOPANTETHEINE
| Macromolecule | Name: 4'-PHOSPHOPANTETHEINE / type: ligand / ID: 2 / Number of copies: 2 / Formula: PNS |
|---|---|
| Molecular weight | Theoretical: 358.348 Da |
| Chemical component information |  ChemComp-PNS: |
-Macromolecule #3: (4S)-2-(2-hydroxyphenyl)-4,5-dihydro-1,3-thiazole-4-carboxylic acid
| Macromolecule | Name: (4S)-2-(2-hydroxyphenyl)-4,5-dihydro-1,3-thiazole-4-carboxylic acid type: ligand / ID: 3 / Number of copies: 1 / Formula: J9F |
|---|---|
| Molecular weight | Theoretical: 223.248 Da |
| Chemical component information | 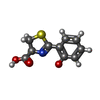 ChemComp-J9F: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: ADENOSINE MONOPHOSPHATE
| Macromolecule | Name: ADENOSINE MONOPHOSPHATE / type: ligand / ID: 5 / Number of copies: 2 / Formula: AMP |
|---|---|
| Molecular weight | Theoretical: 347.221 Da |
| Chemical component information |  ChemComp-AMP: |
-Macromolecule #6: CYSTEINE
| Macromolecule | Name: CYSTEINE / type: ligand / ID: 6 / Number of copies: 2 / Formula: CYS |
|---|---|
| Molecular weight | Theoretical: 121.158 Da |
| Chemical component information | 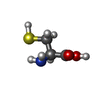 ChemComp-BTC: |
-Macromolecule #7: 2-HYDROXYBENZOIC ACID
| Macromolecule | Name: 2-HYDROXYBENZOIC ACID / type: ligand / ID: 7 / Number of copies: 1 / Formula: SAL |
|---|---|
| Molecular weight | Theoretical: 138.121 Da |
| Chemical component information | 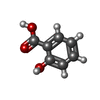 ChemComp-SAL: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
| Details | This sample was homogenous |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 60.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)