[English] 日本語
 Yorodumi
Yorodumi- EMDB-30116: The Cryo-EM Structure of Human Pannexin 1 in the Presence of CBX -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30116 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Cryo-EM Structure of Human Pannexin 1 in the Presence of CBX | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ATP release / Heptameric channel / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationATP transmembrane transporter activity / ATP transport / leak channel activity / Electric Transmission Across Gap Junctions / positive regulation of interleukin-1 alpha production / wide pore channel activity / bleb / monoatomic anion transmembrane transport / monoatomic anion channel activity / gap junction ...ATP transmembrane transporter activity / ATP transport / leak channel activity / Electric Transmission Across Gap Junctions / positive regulation of interleukin-1 alpha production / wide pore channel activity / bleb / monoatomic anion transmembrane transport / monoatomic anion channel activity / gap junction / gap junction channel activity / positive regulation of macrophage cytokine production / Mechanical load activates signaling by PIEZO1 and integrins in osteocytes / oogenesis / response to ATP / The NLRP3 inflammasome / monoatomic cation transport / response to ischemia / positive regulation of interleukin-1 beta production / calcium channel activity / actin filament binding / calcium ion transport / cell-cell signaling / protease binding / scaffold protein binding / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / transmembrane transporter binding / signaling receptor binding / endoplasmic reticulum membrane / structural molecule activity / endoplasmic reticulum / protein-containing complex / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.6 Å | |||||||||
 Authors Authors | Jin QH / Bo Z | |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2020 Journal: Cell Res / Year: 2020Title: Cryo-EM structures of human pannexin 1 channel. Authors: Qiuheng Jin / Bo Zhang / Xiang Zheng / Ningning Li / Lingyi Xu / Yuan Xie / Fangjun Song / Eijaz Ahmed Bhat / Yuan Chen / Ning Gao / Jiangtao Guo / Xiaokang Zhang / Sheng Ye /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30116.map.gz emd_30116.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30116-v30.xml emd-30116-v30.xml emd-30116.xml emd-30116.xml | 10.1 KB 10.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30116.png emd_30116.png | 45 KB | ||
| Filedesc metadata |  emd-30116.cif.gz emd-30116.cif.gz | 5.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30116 http://ftp.pdbj.org/pub/emdb/structures/EMD-30116 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30116 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30116 | HTTPS FTP |
-Related structure data
| Related structure data |  6m68MC  6m66C  6m67C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30116.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30116.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
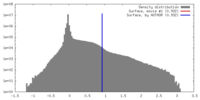
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Pannexin 1
| Entire | Name: Pannexin 1 |
|---|---|
| Components |
|
-Supramolecule #1: Pannexin 1
| Supramolecule | Name: Pannexin 1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Pannexin-1
| Macromolecule | Name: Pannexin-1 / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 49.791629 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAIAQLATEY VFSDFLLKEP TEPKFKGLRL ELAVDKMVTC IAVGLPLLLI SLAFAQEISI GTQISCFSPS SFSWRQAAFV DSYCWAAVQ QKNSLQSESG NLPLWLHKFF PYILLLFAIL LYLPPLFWRF AAAPHICSDL KFIMEELDKV YNRAIKAAKS A RDLDMRDG ...String: MAIAQLATEY VFSDFLLKEP TEPKFKGLRL ELAVDKMVTC IAVGLPLLLI SLAFAQEISI GTQISCFSPS SFSWRQAAFV DSYCWAAVQ QKNSLQSESG NLPLWLHKFF PYILLLFAIL LYLPPLFWRF AAAPHICSDL KFIMEELDKV YNRAIKAAKS A RDLDMRDG ACSVPGVTEN LGQSLWEVSE SHFKYPIVEQ YLKTKKNSNN LIIKYISCRL LTLIIILLAC IYLGYYFSLS SL SDEFVCS IKSGILRNDS TVPDQFQCKL IAVGIFQLLS VINLVVYVLL APVVVYTLFV PFRQKTDVLK VYEILPTFDV LHF KSEGYN DLSLYNLFLE ENISEVKSYK CLKVLENIKS SGQGIDPMLL LTNLGMIKME VVEGKTPMSA EMREEQGNQT AELQ GMNID SETKANNGEK NARQRLLDSS CGSGSHHHHH HHHHH UniProtKB: Pannexin-1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 65.84 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: Ab-inito by cryoSPARC |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.6 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 53207 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)