[English] 日本語
 Yorodumi
Yorodumi- EMDB-28777: Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the acyl... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the acylsulfonamide inhibitor GDC-0310 | |||||||||
 Map data Map data | Density modified map used for model refinements | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationdetection of mechanical stimulus involved in sensory perception / membrane depolarization during action potential / voltage-gated sodium channel complex / cardiac muscle cell action potential involved in contraction / Interaction between L1 and Ankyrins / voltage-gated sodium channel activity / sodium ion transport / behavioral response to pain / Phase 0 - rapid depolarisation / detection of temperature stimulus involved in sensory perception of pain ...detection of mechanical stimulus involved in sensory perception / membrane depolarization during action potential / voltage-gated sodium channel complex / cardiac muscle cell action potential involved in contraction / Interaction between L1 and Ankyrins / voltage-gated sodium channel activity / sodium ion transport / behavioral response to pain / Phase 0 - rapid depolarisation / detection of temperature stimulus involved in sensory perception of pain / neuronal action potential / sodium ion transmembrane transport / sensory perception of pain / post-embryonic development / response to toxic substance / Sensory perception of sweet, bitter, and umami (glutamate) taste / circadian rhythm / inflammatory response / axon / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Kschonsak M / Jao CC / Arthur CP / Rohou AL / Bergeron P / Ortwine D / McKerall SJ / Hackos DH / Deng L / Chen J ...Kschonsak M / Jao CC / Arthur CP / Rohou AL / Bergeron P / Ortwine D / McKerall SJ / Hackos DH / Deng L / Chen J / Sutherlin D / Dragovich PS / Volgraf M / Wright MR / Payandeh J / Ciferri C / Tellis JC | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2023 Journal: Elife / Year: 2023Title: Cryo-EM reveals an unprecedented binding site for Na1.7 inhibitors enabling rational design of potent hybrid inhibitors. Authors: Marc Kschonsak / Christine C Jao / Christopher P Arthur / Alexis L Rohou / Philippe Bergeron / Daniel F Ortwine / Steven J McKerrall / David H Hackos / Lunbin Deng / Jun Chen / Tianbo Li / ...Authors: Marc Kschonsak / Christine C Jao / Christopher P Arthur / Alexis L Rohou / Philippe Bergeron / Daniel F Ortwine / Steven J McKerrall / David H Hackos / Lunbin Deng / Jun Chen / Tianbo Li / Peter S Dragovich / Matthew Volgraf / Matthew R Wright / Jian Payandeh / Claudio Ciferri / John C Tellis /  Abstract: The voltage-gated sodium (Na) channel Na1.7 has been identified as a potential novel analgesic target due to its involvement in human pain syndromes. However, clinically available Na channel-blocking ...The voltage-gated sodium (Na) channel Na1.7 has been identified as a potential novel analgesic target due to its involvement in human pain syndromes. However, clinically available Na channel-blocking drugs are not selective among the nine Na channel subtypes, Na1.1-Na1.9. Moreover, the two currently known classes of Na1.7 subtype-selective inhibitors (aryl- and acylsulfonamides) have undesirable characteristics that may limit their development. To this point understanding of the structure-activity relationships of the acylsulfonamide class of Na1.7 inhibitors, exemplified by the clinical development candidate , has been based solely on a single co-crystal structure of an arylsulfonamide inhibitor bound to voltage-sensing domain 4 (VSD4). To advance inhibitor design targeting the Na1.7 channel, we pursued high-resolution ligand-bound Na1.7-VSD4 structures using cryogenic electron microscopy (cryo-EM). Here, we report that engages the Na1.7-VSD4 through an unexpected binding mode orthogonal to the arylsulfonamide inhibitor class binding pose, which identifies a previously unknown ligand binding site in Na channels. This finding enabled the design of a novel hybrid inhibitor series that bridges the aryl- and acylsulfonamide binding pockets and allows for the generation of molecules with substantially differentiated structures and properties. Overall, our study highlights the power of cryo-EM methods to pursue challenging drug targets using iterative and high-resolution structure-guided inhibitor design. This work also underscores an important role of the membrane bilayer in the optimization of selective Na channel modulators targeting VSD4. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28777.map.gz emd_28777.map.gz | 8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28777-v30.xml emd-28777-v30.xml emd-28777.xml emd-28777.xml | 24 KB 24 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28777.png emd_28777.png | 79.1 KB | ||
| Others |  emd_28777_additional_1.map.gz emd_28777_additional_1.map.gz emd_28777_half_map_1.map.gz emd_28777_half_map_1.map.gz emd_28777_half_map_2.map.gz emd_28777_half_map_2.map.gz | 226.1 MB 46.6 MB 46.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28777 http://ftp.pdbj.org/pub/emdb/structures/EMD-28777 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28777 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28777 | HTTPS FTP |
-Validation report
| Summary document |  emd_28777_validation.pdf.gz emd_28777_validation.pdf.gz | 800.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28777_full_validation.pdf.gz emd_28777_full_validation.pdf.gz | 800.1 KB | Display | |
| Data in XML |  emd_28777_validation.xml.gz emd_28777_validation.xml.gz | 13.7 KB | Display | |
| Data in CIF |  emd_28777_validation.cif.gz emd_28777_validation.cif.gz | 15.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28777 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28777 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28777 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28777 | HTTPS FTP |
-Related structure data
| Related structure data |  8f0qMC  8f0pC  8f0rC  8f0sC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28777.map.gz / Format: CCP4 / Size: 8.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28777.map.gz / Format: CCP4 / Size: 8.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density modified map used for model refinements | ||||||||||||||||||||||||||||||||||||
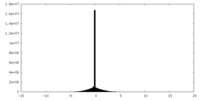
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.838 Å | ||||||||||||||||||||||||||||||||||||
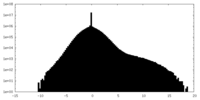
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: non-modified, full map
| File | emd_28777_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | non-modified, full map | ||||||||||||
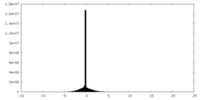
| Projections & Slices |
| ||||||||||||
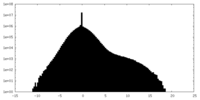
| Density Histograms |
-Half map: half-map1
| File | emd_28777_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half-map2
| File | emd_28777_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the acyl...
| Entire | Name: Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the acylsulfonamide inhibitor GDC-0310 |
|---|---|
| Components |
|
-Supramolecule #1: Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the acyl...
| Supramolecule | Name: Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the acylsulfonamide inhibitor GDC-0310 type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1 Details: Chimeric construct of human Nav1.7 VSD4 and the NavPaS channel from American cockroach Periplaneta americana |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Sodium channel protein PaFPC1,Sodium channel protein type 9 subun...
| Macromolecule | Name: Sodium channel protein PaFPC1,Sodium channel protein type 9 subunit alpha chimera type: protein_or_peptide / ID: 1 Details: Chimeric construct of human Nav1.7 VSD4 and the NavPaS channel from American cockroach Periplaneta americana Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 184.481906 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MWSHPQFEKG GGSGGGSGGS AWSHPQFEKG GSGGDYKDDD DKGGSGGDYK DDDDKMADNS PLIREERQRL FRPYTRAMLT APSAQPAKE NGKTEENKDN SRDKGRGANK DRDGSAHPDQ ALEQGSRLPA RMRNIFPAEL ASTPLEDFDP FYKNKKTFVV V TKAGDIFR ...String: MWSHPQFEKG GGSGGGSGGS AWSHPQFEKG GSGGDYKDDD DKGGSGGDYK DDDDKMADNS PLIREERQRL FRPYTRAMLT APSAQPAKE NGKTEENKDN SRDKGRGANK DRDGSAHPDQ ALEQGSRLPA RMRNIFPAEL ASTPLEDFDP FYKNKKTFVV V TKAGDIFR FSGEKSLWML DPFTPIRRVA ISTMVQPIFS YFIMITILIH CIFMIMPATQ TTYILELVFL SIYTIEVVVK VL ARGFILH PFAYLRDPWN WLDFLVTLIG YITLVVDLGH LYALRAFRVL RSWRTVTIVP GWRTIVDALS LSITSLKDLV LLL LFSLSV FALIGLQLFM GNLKHKCVKH FPADGSWGNF TDERWFNYTS NSSHWYIPDD WIEYPLCGNS SGAGMCPPGY TCLQ GYGGN PNYGYTSFDT FGWAFLSVFR LVTLDYWEDL YQLALRSAGP WHILFFIIVV FYGTFCFLNF ILAVVVMSYT HMVKR ADEE KAAERELKKE KKAASVANNT ANGQEQTTIE MNGDEAVVID NNDQAARQQS DPETPAPSVT QRLTDFLCVW DCCVPW QKL QGAIGAVVLS PFFELFIAVI IVLNITFMAL DHHDMNIEFE RILRTGNYIF TSIYIVEAVL KIIALSPKFY FKDSWNV FD FIIVVFAILE LGLEGVQGLS VFRSFRLLRV FRLAKFWPTL NNFMSVMTKS YGAFVNVMYV MFLLLFIFAI IGMQLFGM N YIDNMERFPD GDLPRWNFTD FLHSFMIVFR ALCGEWIESM WDCMLVGDWS CIPFFVAVFF VGNLVILNLL IALLLNNYG SFCTSPTSDE EDSKDEDALA QIVRIFKRFK PNLNAVKLSP MKPDSEDIVE SQEIQGNNIA DAEDVLAGEF PPDCCCNAFY KCFPSRPAR DSSVQRMWSN IRRVCFLLAK NKYFQKFVTA VLVITSVLLA LEDIYLPQRP VLVNITLYVD YVLTAFFVIE M IIMLFAVG FKKYFTSKWY WLDFIVVVAY LLNFVLMCAG IEALQTLRLL RVFRLFRPLS KVNGMQVVTS TLVEAVPHIF NV ILVGIFF WLVFAIMGVQ LFAGKFYKCV DENSTVLSHE ITMDRNDCLH ENYTWENSPM NFDHVGNAYL SLLQVATFKG WLQ IMNDAI DSREVHKQPI RETNIYMYLY FIFFIVFGSF FILKLFVCIL IDIFRQQRRK AEGLSATDSR TQLIYRRAVM RTMS AKPVK RIPKPGNKIQ GCIFDLVTNQ AFDISIMVLI CLNMVTMMVE KEGQSQHMTE VLYWINVVFI ILFTGECVLK LISLR HYYF TVGWNIFDFV VVIISIVGMF LADLIETYFV SPTLFRVIRL ARIGRILRLV KGAKGIRLLL LALRKALRTL FNVSFL LFV IMFVYAVFGM EFFMHIRDAG AIDDVYNFKT FGQSIILLFQ LATSAGWDGV YFAIANEEDC RAPDHELGYP GNCGSRA LG IAYLVSYLII TCLVVINMYA AVILDYVLEV YEDSKEGLTD DDYDMFFEVW QQFDPEATQY IRYDQLSELL EALQPPLQ V QKPNKYKILS MNIPICKDDH IFYKDVLEAL VKDVFSRRGS PVEAGDVQAP NVDEAEYKPV SSTLQRQREE YCVRLIQNA WRKHKQQN |
-Macromolecule #3: beta-D-mannopyranose
| Macromolecule | Name: beta-D-mannopyranose / type: ligand / ID: 3 / Number of copies: 1 / Formula: BMA |
|---|---|
| Molecular weight | Theoretical: 180.156 Da |
| Chemical component information |  ChemComp-BMA: |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #5: 5-cyclopropyl-4-({1-[(1S)-1-(3,5-dichlorophenyl)ethyl]piperidin-4...
| Macromolecule | Name: 5-cyclopropyl-4-({1-[(1S)-1-(3,5-dichlorophenyl)ethyl]piperidin-4-yl}methoxy)-2-fluoro-N-(methanesulfonyl)benzamide type: ligand / ID: 5 / Number of copies: 1 / Formula: X7R |
|---|---|
| Molecular weight | Theoretical: 543.478 Da |
| Chemical component information | 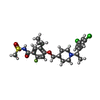 ChemComp-X7R: |
-Macromolecule #6: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
| Macromolecule | Name: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine / type: ligand / ID: 6 / Number of copies: 5 / Formula: PEE |
|---|---|
| Molecular weight | Theoretical: 744.034 Da |
| Chemical component information |  ChemComp-PEE: |
-Macromolecule #7: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 7 / Number of copies: 1 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 78 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: UltrAuFoil R0./1 / Material: GOLD / Mesh: 300 Details: Grids were incubated with a thiol reactive, self-assembling reaction mixture of 4mM monothiolalkane(C11)PEG6-OH (11-mercaptoundecyl) hexaethyleneglycol (SPT-0011P6, SensoPath Technologies, ...Details: Grids were incubated with a thiol reactive, self-assembling reaction mixture of 4mM monothiolalkane(C11)PEG6-OH (11-mercaptoundecyl) hexaethyleneglycol (SPT-0011P6, SensoPath Technologies, Inc., Bozeman, MT). Grids were incubated with this self-assembled monolayer (SAM) solution for 24 hours and afterwards rinsed with EtOH. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | The sample was reconstituted into lipid nanodiscs (MSP1E3D1 in 3POPC:1POPE:1POPG) and was monodisperse. The sample was crosslinked with 0.05% glutaraldehyde for 10 minutes at RT, then quenched with 1M Tris pH7.0. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 3.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8f0q: |
 Movie
Movie Controller
Controller










 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)