[English] 日本語
 Yorodumi
Yorodumi- EMDB-27939: Structure of the human ACE2 receptor in complex with antibody Fab... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the human ACE2 receptor in complex with antibody Fab fragment, 05B04 | |||||||||
 Map data Map data | main map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ACE2 / antibody / SARS-CoV-2 / inhibition / ANTIVIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of amino acid transport / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / positive regulation of gap junction assembly / regulation of systemic arterial blood pressure by renin-angiotensin / tryptophan transport / regulation of cardiac conduction / maternal process involved in female pregnancy ...positive regulation of amino acid transport / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / positive regulation of gap junction assembly / regulation of systemic arterial blood pressure by renin-angiotensin / tryptophan transport / regulation of cardiac conduction / maternal process involved in female pregnancy / peptidyl-dipeptidase activity / regulation of vasoconstriction / transporter activator activity / Metabolism of Angiotensinogen to Angiotensins / carboxypeptidase activity / angiotensin maturation / viral life cycle / Attachment and Entry / receptor-mediated endocytosis of virus by host cell / metallocarboxypeptidase activity / positive regulation of cardiac muscle contraction / regulation of cytokine production / blood vessel diameter maintenance / negative regulation of smooth muscle cell proliferation / brush border membrane / negative regulation of ERK1 and ERK2 cascade / positive regulation of reactive oxygen species metabolic process / metallopeptidase activity / endocytic vesicle membrane / regulation of cell population proliferation / virus receptor activity / regulation of inflammatory response / endopeptidase activity / viral translation / Induction of Cell-Cell Fusion / Potential therapeutics for SARS / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / receptor-mediated virion attachment to host cell / cilium / apical plasma membrane / membrane raft / endoplasmic reticulum lumen / symbiont entry into host cell / cell surface / negative regulation of transcription by RNA polymerase II / extracellular space / extracellular exosome / extracellular region / zinc ion binding / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
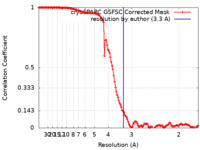
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Barnes CO | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Biorxiv / Year: 2022 Journal: Biorxiv / Year: 2022Title: Human anti-ACE2 monoclonal antibodies as pan-sarbecovirus prophylactic agents Authors: Zhang F / Jenkins J / de Carvalho RV / Nakandakari-Higa S / Chen T / Abernathy ME / Nyakatura E / Andrew D / Lebedeva I / Lorenz IC / Hoffmann HH / Rice CM / Victora GD / Barnes CO / Haziioannou T / Bieniasz PD | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27939.map.gz emd_27939.map.gz | 202.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27939-v30.xml emd-27939-v30.xml emd-27939.xml emd-27939.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27939_fsc.xml emd_27939_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_27939.png emd_27939.png | 69.8 KB | ||
| Masks |  emd_27939_msk_1.map emd_27939_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-27939.cif.gz emd-27939.cif.gz | 7.1 KB | ||
| Others |  emd_27939_additional_1.map.gz emd_27939_additional_1.map.gz emd_27939_half_map_1.map.gz emd_27939_half_map_1.map.gz emd_27939_half_map_2.map.gz emd_27939_half_map_2.map.gz | 106.7 MB 200.6 MB 200.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27939 http://ftp.pdbj.org/pub/emdb/structures/EMD-27939 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27939 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27939 | HTTPS FTP |
-Related structure data
| Related structure data |  8e7mMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27939.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27939.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.867 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27939_msk_1.map emd_27939_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: unsharpened map
| File | emd_27939_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_27939_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_27939_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : hACE2 + 05B04 Fab fragment
| Entire | Name: hACE2 + 05B04 Fab fragment |
|---|---|
| Components |
|
-Supramolecule #1: hACE2 + 05B04 Fab fragment
| Supramolecule | Name: hACE2 + 05B04 Fab fragment / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: Complex between soluble ACE2 bound to 05B04 Fab fragment |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 100 KDa |
-Macromolecule #1: Angiotensin-converting enzyme 2
| Macromolecule | Name: Angiotensin-converting enzyme 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: angiotensin-converting enzyme 2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 72.306203 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSSSSWLLLS LVAVTAAQST IEEQAKTFLD KFNHEAEDLF YQSSLASWNY NTNITEENVQ NMNNAGDKWS AFLKEQSTLA QMYPLQEIQ NLTVKLQLQA LQQNGSSVLS EDKSKRLNTI LNTMSTIYST GKVCNPDNPQ ECLLLEPGLN EIMANSLDYN E RLWAWESW ...String: MSSSSWLLLS LVAVTAAQST IEEQAKTFLD KFNHEAEDLF YQSSLASWNY NTNITEENVQ NMNNAGDKWS AFLKEQSTLA QMYPLQEIQ NLTVKLQLQA LQQNGSSVLS EDKSKRLNTI LNTMSTIYST GKVCNPDNPQ ECLLLEPGLN EIMANSLDYN E RLWAWESW RSEVGKQLRP LYEEYVVLKN EMARANHYED YGDYWRGDYE VNGVDGYDYS RGQLIEDVEH TFEEIKPLYE HL HAYVRAK LMNAYPSYIS PIGCLPAHLL GDMWGRFWTN LYSLTVPFGQ KPNIDVTDAM VDQAWDAQRI FKEAEKFFVS VGL PNMTQG FWENSMLTDP GNVQKAVCHP TAWDLGKGDF RILMCTKVTM DDFLTAHHEM GHIQYDMAYA AQPFLLRNGA NEGF HEAVG EIMSLSAATP KHLKSIGLLS PDFQEDNETE INFLLKQALT IVGTLPFTYM LEKWRWMVFK GEIPKDQWMK KWWEM KREI VGVVEPVPHD ETYCDPASLF HVSNDYSFIR YYTRTLYQFQ FQEALCQAAK HEGPLHKCDI SNSTEAGQKL FNMLRL GKS EPWTLALENV VGAKNMNVRP LLNYFEPLFT WLKDQNKNSF VGWSTDWSPY ADGSGHHHHH HHH UniProtKB: Angiotensin-converting enzyme 2 |
-Macromolecule #2: 05B04 Heavy Chain
| Macromolecule | Name: 05B04 Heavy Chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.948818 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLLESGGG LVQPGGSLRL SCAASGFTFS SYAMSWVRQA PGKGLEWVSG ISGSGDRTYY PDSVKGRFTI SRDNSKNTLY LQMNSLRAE DTAVYYCAKD WAMVGADAFD IWGQGTMVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL VKDYFPEPVT V SWNSGALT ...String: EVQLLESGGG LVQPGGSLRL SCAASGFTFS SYAMSWVRQA PGKGLEWVSG ISGSGDRTYY PDSVKGRFTI SRDNSKNTLY LQMNSLRAE DTAVYYCAKD WAMVGADAFD IWGQGTMVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL VKDYFPEPVT V SWNSGALT SGVHTFPAVL QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKK VEPKSCDKT |
-Macromolecule #3: 05B04 Light Chain
| Macromolecule | Name: 05B04 Light Chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.549191 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSA MSASVGDRVT ITCRASQGIS NYFAWFQQKP GKVPKRLIYA ASSLQSGVPS RFSGSGSGTE FTLTISNLQP EDFATYYCL QHSYYPYTFG QGTKLEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...String: DIQMTQSPSA MSASVGDRVT ITCRASQGIS NYFAWFQQKP GKVPKRLIYA ASSLQSGVPS RFSGSGSGTE FTLTISNLQP EDFATYYCL QHSYYPYTFG QGTKLEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.0 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 6599 / Average exposure time: 2.6 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)