[English] 日本語
 Yorodumi
Yorodumi- EMDB-27787: Cryo-EM structure of 337 Fab in complex with recombinant shortene... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of 337 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP) | |||||||||
 Map data Map data | Cryo-EM structure of 337 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | malaria antibody / PfCSP / IMMUNE SYSTEM | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Martin GM / Ward AB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Affinity-matured homotypic interactions induce spectrum of PfCSP structures that influence protection from malaria infection. Authors: Gregory M Martin / Jonathan L Torres / Tossapol Pholcharee / David Oyen / Yevel Flores-Garcia / Grace Gibson / Re'em Moskovitz / Nathan Beutler / Diana D Jung / Jeffrey Copps / Wen-Hsin Lee ...Authors: Gregory M Martin / Jonathan L Torres / Tossapol Pholcharee / David Oyen / Yevel Flores-Garcia / Grace Gibson / Re'em Moskovitz / Nathan Beutler / Diana D Jung / Jeffrey Copps / Wen-Hsin Lee / Gonzalo Gonzalez-Paez / Daniel Emerling / Randall S MacGill / Emily Locke / C Richter King / Fidel Zavala / Ian A Wilson / Andrew B Ward /   Abstract: The generation of high-quality antibody responses to Plasmodium falciparum (Pf) circumsporozoite protein (PfCSP), the primary surface antigen of Pf sporozoites, is paramount to the development of an ...The generation of high-quality antibody responses to Plasmodium falciparum (Pf) circumsporozoite protein (PfCSP), the primary surface antigen of Pf sporozoites, is paramount to the development of an effective malaria vaccine. Here we present an in-depth structural and functional analysis of a panel of potent antibodies encoded by the immunoglobulin heavy chain variable (IGHV) gene IGHV3-33, which is among the most prevalent and potent antibody families induced in the anti-PfCSP immune response and targets the Asn-Ala-Asn-Pro (NANP) repeat region. Cryo-electron microscopy (cryo-EM) reveals a remarkable spectrum of helical antibody-PfCSP structures stabilized by homotypic interactions between tightly packed fragments antigen binding (Fabs), many of which correlate with somatic hypermutation. We demonstrate a key role of these mutated homotypic contacts for high avidity binding to PfCSP and in protection from Pf malaria infection. Together, these data emphasize the importance of anti-homotypic affinity maturation in the frequent selection of IGHV3-33 antibodies and highlight key features underlying the potent protection of this antibody family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27787.map.gz emd_27787.map.gz | 37.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27787-v30.xml emd-27787-v30.xml emd-27787.xml emd-27787.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27787.png emd_27787.png | 162.4 KB | ||
| Filedesc metadata |  emd-27787.cif.gz emd-27787.cif.gz | 6.5 KB | ||
| Others |  emd_27787_half_map_1.map.gz emd_27787_half_map_1.map.gz emd_27787_half_map_2.map.gz emd_27787_half_map_2.map.gz | 37.1 MB 37.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27787 http://ftp.pdbj.org/pub/emdb/structures/EMD-27787 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27787 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27787 | HTTPS FTP |
-Related structure data
| Related structure data |  8dz3MC  8dytC  8dywC  8dyxC  8dyyC  8dz4C  8dz5C  8ekfC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27787.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27787.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of 337 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.026 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Cryo-EM structure of 337 Fab in complex with...
| File | emd_27787_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of 337 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP) | ||||||||||||
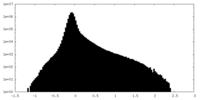
| Projections & Slices |
| ||||||||||||
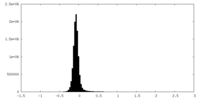
| Density Histograms |
-Half map: Cryo-EM structure of 337 Fab in complex with...
| File | emd_27787_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of 337 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP) | ||||||||||||
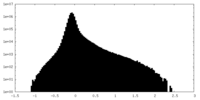
| Projections & Slices |
| ||||||||||||
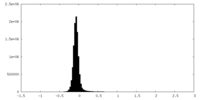
| Density Histograms |
- Sample components
Sample components
-Entire : 337 Fab - rsCSP complex
| Entire | Name: 337 Fab - rsCSP complex |
|---|---|
| Components |
|
-Supramolecule #1: 337 Fab - rsCSP complex
| Supramolecule | Name: 337 Fab - rsCSP complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Circumsporozoite protein
| Supramolecule | Name: Circumsporozoite protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: 337 Fab heavy chain, 337 Fab light chain
| Supramolecule | Name: 337 Fab heavy chain, 337 Fab light chain / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Circumsporozoite protein
| Macromolecule | Name: Circumsporozoite protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30.232156 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: YGSSSNTRVL NELNYDNAGT NLYNELEMNY YGKQENWYSL KKNSRSLGEN DDGNNEDNEK LRKPKHKKLK QPADGNPDPN ANPNVDPNA NPNVDPNANP NVDPNANPNA NPNANPNANP NANPNANPNA NPNANPNANP NANPNANPNA NPNANPNANP N ANPNANPN ...String: YGSSSNTRVL NELNYDNAGT NLYNELEMNY YGKQENWYSL KKNSRSLGEN DDGNNEDNEK LRKPKHKKLK QPADGNPDPN ANPNVDPNA NPNVDPNANP NVDPNANPNA NPNANPNANP NANPNANPNA NPNANPNANP NANPNANPNA NPNANPNANP N ANPNANPN KNNQGNGQGH NMPNDPNRNV DENANANSAV KNNNNEEPSD KHIKEYLNKI QNSLSTEWSP CSVTCGNGIQ VR IKPGSAN KPKDELDYAN DIEKKICKME KCSSVFNVVN S |
-Macromolecule #2: 337 Fab heavy chain
| Macromolecule | Name: 337 Fab heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.31923 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLVESGGG VVQPGRSLRL SCAASGFSFS TYGMHWVRQA PGKGLEWVAL IWHDGSKKFY SESVRGRFTI SRDNSMDTLY LQMSSLRAE DTAVYYCARD GADYFDSTGG AFDIWGQGTM VIVSSASTKG PSVFPLAPSS KSTSGGTAAL GCLVKDYFPE P VTVSWNSG ...String: QVQLVESGGG VVQPGRSLRL SCAASGFSFS TYGMHWVRQA PGKGLEWVAL IWHDGSKKFY SESVRGRFTI SRDNSMDTLY LQMSSLRAE DTAVYYCARD GADYFDSTGG AFDIWGQGTM VIVSSASTKG PSVFPLAPSS KSTSGGTAAL GCLVKDYFPE P VTVSWNSG ALTSGVHTFP AVLQSSGLYS LSSVVTVPSS SLGTQTYICN VNHKPSNTKV DKKVEPKSCD |
-Macromolecule #3: 337 Fab light chain
| Macromolecule | Name: 337 Fab light chain / type: protein_or_peptide / ID: 3 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.321887 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EIVMTQSPAT LSVSPGERAT LSCRASQSVS SNLAWYQQKP GQAPRLLIYG ASTRATGIPA RFSGSGSGTE FTLTISSLQS EDFATYYCQ QYKTWWTFGQ GTKVEIKRTV AAPSVFIFPP SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ E SVTEQDSK ...String: EIVMTQSPAT LSVSPGERAT LSCRASQSVS SNLAWYQQKP GQAPRLLIYG ASTRATGIPA RFSGSGSGTE FTLTISSLQS EDFATYYCQ QYKTWWTFGQ GTKVEIKRTV AAPSVFIFPP SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ E SVTEQDSK DSTYSLSSTL TLSKADYEKH KVYACEVTHQ GLSSPVTKSF NRGEC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)