[English] 日本語
 Yorodumi
Yorodumi- EMDB-27750: Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
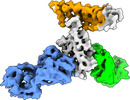
| Title | Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR16m, and two antibody Fabs, S309 and CR3022 | |||||||||
 Map data Map data | SARS-CoV-2 RBD structure in complex with DARPin, SR16m, and antibody Fabs, S309 and CR3022 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DARPins / Anti-SARS-CoV-2 / therapeutics / COVID-19 / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
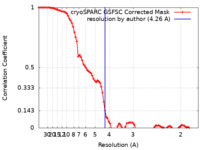
| Method | single particle reconstruction / cryo EM / Resolution: 4.26 Å | |||||||||
 Authors Authors | Kwon YD / Gorman J / Kwong PD | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2023 Journal: Nat Chem Biol / Year: 2023Title: A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins. Authors: Vikas Chonira / Young D Kwon / Jason Gorman / James Brett Case / Zhiqiang Ku / Rudo Simeon / Ryan G Casner / Darcy R Harris / Adam S Olia / Tyler Stephens / Lawrence Shapiro / Michael F ...Authors: Vikas Chonira / Young D Kwon / Jason Gorman / James Brett Case / Zhiqiang Ku / Rudo Simeon / Ryan G Casner / Darcy R Harris / Adam S Olia / Tyler Stephens / Lawrence Shapiro / Michael F Bender / Hannah Boyd / I-Ting Teng / Yaroslav Tsybovsky / Florian Krammer / Ningyan Zhang / Michael S Diamond / Peter D Kwong / Zhiqiang An / Zhilei Chen /  Abstract: We report the engineering and selection of two synthetic proteins-FSR16m and FSR22-for the possible treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. FSR16m and ...We report the engineering and selection of two synthetic proteins-FSR16m and FSR22-for the possible treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. FSR16m and FSR22 are trimeric proteins composed of DARPin SR16m or SR22 fused with a T4 foldon. Despite selection by a spike protein from a now historical SARS-CoV-2 strain, FSR16m and FSR22 exhibit broad-spectrum neutralization of SARS-CoV-2 strains, inhibiting authentic B.1.351, B.1.617.2 and BA.1.1 viruses, with respective IC values of 3.4, 2.2 and 7.4 ng ml for FSR16m. Cryo-EM structures revealed that these DARPins recognize a region of the receptor-binding domain (residues 456, 475, 486, 487 and 489) overlapping a critical portion of the angiotensin-converting enzyme 2 (ACE2)-binding surface. K18-hACE2 transgenic mice inoculated with B.1.617.2 and receiving intranasally administered FSR16m showed less weight loss and 10-100-fold lower viral burden in upper and lower respiratory tracts. The strong and broad neutralization potency makes FSR16m and FSR22 promising candidates for the prevention and treatment of infection by SARS-CoV-2. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27750.map.gz emd_27750.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27750-v30.xml emd-27750-v30.xml emd-27750.xml emd-27750.xml | 22.3 KB 22.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27750_fsc.xml emd_27750_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_27750.png emd_27750.png | 104.8 KB | ||
| Filedesc metadata |  emd-27750.cif.gz emd-27750.cif.gz | 6.7 KB | ||
| Others |  emd_27750_half_map_1.map.gz emd_27750_half_map_1.map.gz emd_27750_half_map_2.map.gz emd_27750_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27750 http://ftp.pdbj.org/pub/emdb/structures/EMD-27750 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27750 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27750 | HTTPS FTP |
-Related structure data
| Related structure data |  8dw3MC  7tyzC  7tz0C  8dw2C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27750.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27750.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SARS-CoV-2 RBD structure in complex with DARPin, SR16m, and antibody Fabs, S309 and CR3022 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.873 Å | ||||||||||||||||||||||||||||||||||||
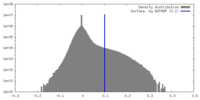
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: SARS-CoV-2 RBD structure in complex with DARPin, SR16m,...
| File | emd_27750_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SARS-CoV-2 RBD structure in complex with DARPin, SR16m, and antibody Fabs, S309 and CR3022 | ||||||||||||
| Projections & Slices |
| ||||||||||||
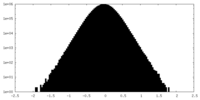
| Density Histograms |
-Half map: SARS-CoV-2 RBD structure in complex with DARPin, SR16m,...
| File | emd_27750_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SARS-CoV-2 RBD structure in complex with DARPin, SR16m, and antibody Fabs, S309 and CR3022 | ||||||||||||
| Projections & Slices |
| ||||||||||||
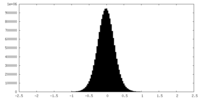
| Density Histograms |
- Sample components
Sample components
-Entire : DARPin SR16m in complex with SARS-CoV-2 RBD and antibody Fabs, S3...
| Entire | Name: DARPin SR16m in complex with SARS-CoV-2 RBD and antibody Fabs, S309 and CR3022 |
|---|---|
| Components |
|
-Supramolecule #1: DARPin SR16m in complex with SARS-CoV-2 RBD and antibody Fabs, S3...
| Supramolecule | Name: DARPin SR16m in complex with SARS-CoV-2 RBD and antibody Fabs, S309 and CR3022 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: DARPin SR16m in complex with SARS-CoV-2 RBD and antibody Fabs, S309 and CR3022 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Anti-SARS-CoV-2 DARPin SR16m
| Macromolecule | Name: Anti-SARS-CoV-2 DARPin SR16m / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 16.793023 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GKKLLEAARA GQDDEVRILM ANGADVNALD LFGVTPLHLA AERGHLEIVE VLLKNGADVN AGDAFGRTPL HLAALGGHLE IVEVLLKNG ADVNACDLYG VTPLHLAAGL GHLEIVEVLL KNGADVNAQD KFGKTAFDIS IDNGNEDLAE ILQSSSKLAA A L |
-Macromolecule #2: Spike protein S1
| Macromolecule | Name: Spike protein S1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.100758 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: PNITNLCPFG EVFNATRFAS VYAWNRKRIS NCVADYSVLY NSASFSTFKC YGVSPTKLND LCFTNVYADS FVIRGDEVRQ IAPGQTGKI ADYNYKLPDD FTGCVIAWNS NNLDSKVGGN YNYLYRLFRK SNLKPFERDI STEIYQAGST PCNGVEGFNC Y FPLQSYGF ...String: PNITNLCPFG EVFNATRFAS VYAWNRKRIS NCVADYSVLY NSASFSTFKC YGVSPTKLND LCFTNVYADS FVIRGDEVRQ IAPGQTGKI ADYNYKLPDD FTGCVIAWNS NNLDSKVGGN YNYLYRLFRK SNLKPFERDI STEIYQAGST PCNGVEGFNC Y FPLQSYGF QPTNGVGYQP YRVVVLSFEL LHAPATVCG UniProtKB: Spike glycoprotein |
-Macromolecule #3: Antibody S309 light chain
| Macromolecule | Name: Antibody S309 light chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.204697 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPGT LSLSPGERAT LSCRASQTVS STSLAWYQQK PGQAPRLLIY GASSRATGIP DRFSGSGSGT DFTLTISRLE PEDFAVYYC QQHDTSLTFG GGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...String: EIVLTQSPGT LSLSPGERAT LSCRASQTVS STSLAWYQQK PGQAPRLLIY GASSRATGIP DRFSGSGSGT DFTLTISRLE PEDFAVYYC QQHDTSLTFG GGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC |
-Macromolecule #4: antibody S309 heavy chain
| Macromolecule | Name: antibody S309 heavy chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.573471 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE VKKPGASVKV SCKASGYPFT SYGISWVRQA PGQGLEWMGW ISTYNGNTNY AQKFQGRVTM TTDTSTTTGY MELRRLRSD DTAVYYCARD YTRGAWFGES LIGGFDNWGQ GTLVTVSSAS TKGPSVFPLA PSSKSTSGGT AALGCLVKDY F PEPVTVSW ...String: QVQLVQSGAE VKKPGASVKV SCKASGYPFT SYGISWVRQA PGQGLEWMGW ISTYNGNTNY AQKFQGRVTM TTDTSTTTGY MELRRLRSD DTAVYYCARD YTRGAWFGES LIGGFDNWGQ GTLVTVSSAS TKGPSVFPLA PSSKSTSGGT AALGCLVKDY F PEPVTVSW NSGALTSGVH TFPAVLQSSG LYSLSSVVTV PSSSLGTQTY ICNVNHKPSN TKVDKKVEPK SC |
-Macromolecule #5: antibody CR3022 heavy chain
| Macromolecule | Name: antibody CR3022 heavy chain / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.382369 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TQMQLVQSGT EVKKPGESLK ISCKGSGYGF ITYWIGWVRQ MPGKGLEWMG IIYPGDSETR YSPSFQGQVT ISADKSINTA YLQWSSLKA SDTAIYYCAG GSGISTPMDV WGQGTTVTVA STKGPSVFPL APSSKSTSGG TAALGCLVKD YFPEPVTVSW N SGALTSGV ...String: TQMQLVQSGT EVKKPGESLK ISCKGSGYGF ITYWIGWVRQ MPGKGLEWMG IIYPGDSETR YSPSFQGQVT ISADKSINTA YLQWSSLKA SDTAIYYCAG GSGISTPMDV WGQGTTVTVA STKGPSVFPL APSSKSTSGG TAALGCLVKD YFPEPVTVSW N SGALTSGV HTFPAVLQSS GLYSLSSVVT VPSSSLGTQT YICNVNHKPS NTKVDKKVEP KSC |
-Macromolecule #6: antibody CR3022 light chain
| Macromolecule | Name: antibody CR3022 light chain / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.289885 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQLTQSPDS LAVSLGERAT INCKSSQSVL YSSINKNYLA WYQQKPGQPP KLLIYWASTR ESGVPDRFSG SGSGTDFTLT ISSLQAEDV AVYYCQQYYS TPYTFGQGTK VEIKRTVAAP SVFIFPPSDE QLKSGTASVV CLLNNFYPRE AKVQWKVDNA L QSGNSQES ...String: DIQLTQSPDS LAVSLGERAT INCKSSQSVL YSSINKNYLA WYQQKPGQPP KLLIYWASTR ESGVPDRFSG SGSGTDFTLT ISSLQAEDV AVYYCQQYYS TPYTFGQGTK VEIKRTVAAP SVFIFPPSDE QLKSGTASVV CLLNNFYPRE AKVQWKVDNA L QSGNSQES VTEQDSKDST YSLSSTLTLS KADYEKHKVY ACEVTHQGLS SPVTKSFNRG EC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 10 mM HEPES, 7.4, 150 mM NaCl |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.25 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 134 / Target criteria: CC |
| Output model |  PDB-8dw3: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)