[English] 日本語
 Yorodumi
Yorodumi- EMDB-27441: CryoEM structure of Influenza A virus A/Melbourne/1/1946 (H1N1) h... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Influenza A virus A/Melbourne/1/1946 (H1N1) hemagglutinin bound to CR6261 Fab | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NIAID hemagglutinin stalk binding antibody / Structural Genomics / Seattle Structural Genomics Center for Infectious Disease / SSGCID / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Biological species |   Influenza A virus / Influenza A virus /  Homo sapiens (human) Homo sapiens (human) | |||||||||
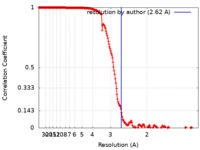
| Method | single particle reconstruction / cryo EM / Resolution: 2.62 Å | |||||||||
 Authors Authors | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To be published Journal: To be publishedTitle: CryoEM structure of Influenza A virus A/Melbourne/1/1946 (H1N1) hemagglutinin bound to CR6261 Fab Authors: Yang M / Edwards TE / Horanyi PS / Lorimer DD | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27441.map.gz emd_27441.map.gz | 624.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27441-v30.xml emd-27441-v30.xml emd-27441.xml emd-27441.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27441_fsc.xml emd_27441_fsc.xml | 19.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_27441.png emd_27441.png | 56.6 KB | ||
| Filedesc metadata |  emd-27441.cif.gz emd-27441.cif.gz | 6.8 KB | ||
| Others |  emd_27441_half_map_1.map.gz emd_27441_half_map_1.map.gz emd_27441_half_map_2.map.gz emd_27441_half_map_2.map.gz | 614.8 MB 614.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27441 http://ftp.pdbj.org/pub/emdb/structures/EMD-27441 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27441 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27441 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27441.map.gz / Format: CCP4 / Size: 662.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27441.map.gz / Format: CCP4 / Size: 662.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.675 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_27441_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
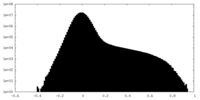
| Projections & Slices |
| ||||||||||||
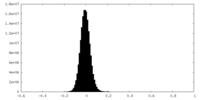
| Density Histograms |
-Half map: #2
| File | emd_27441_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
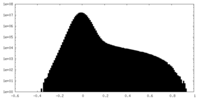
| Projections & Slices |
| ||||||||||||
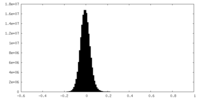
| Density Histograms |
- Sample components
Sample components
-Entire : CryoEM structure of Influenza A virus A/Melbourne/1/1946 (H1N1) h...
| Entire | Name: CryoEM structure of Influenza A virus A/Melbourne/1/1946 (H1N1) hemagglutinin bound to CR6261 Fab |
|---|---|
| Components |
|
-Supramolecule #1: CryoEM structure of Influenza A virus A/Melbourne/1/1946 (H1N1) h...
| Supramolecule | Name: CryoEM structure of Influenza A virus A/Melbourne/1/1946 (H1N1) hemagglutinin bound to CR6261 Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Molecular weight | Theoretical: 450 KDa |
-Macromolecule #1: Hemagglutinin HA1 chain
| Macromolecule | Name: Hemagglutinin HA1 chain / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Influenza A virus / Strain: A/Melbourne/1/1946(H1N1) Influenza A virus / Strain: A/Melbourne/1/1946(H1N1) |
| Molecular weight | Theoretical: 40.34752 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MVLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFAGS DTICIGYHAN NSTDTVDTVL EKNVTVTHSV NLLEDSHNGK LCRLKGIAP LQLGKCNIAG WILGNPECDS LLPASSWSYI VETPNSKNGI CYPGDFIDYE ELREQLSSVS SFERFEIFPK E SSWPKHST ...String: MVLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFAGS DTICIGYHAN NSTDTVDTVL EKNVTVTHSV NLLEDSHNGK LCRLKGIAP LQLGKCNIAG WILGNPECDS LLPASSWSYI VETPNSKNGI CYPGDFIDYE ELREQLSSVS SFERFEIFPK E SSWPKHST TKGVTAACSH AGKSSFYRNL LWLTKKEDSY PKLSNSYVNK KGKEVLVLWG VHHPSSSKEQ QTLYQNENAY VS VVSSNYN RRFIPEIAER PEVKDQAGRI NYYWTLLEPG DTIIFEANGN LVAPWYAFAL SRGFGSGIIT SNASMHECNT KCQ TPQGAI NSSLPFQNIH PVTIGECPKY VKSAKLRMVT GLRNIPSI |
-Macromolecule #2: Hemagglutinin HA2 chain
| Macromolecule | Name: Hemagglutinin HA2 chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Influenza A virus / Strain: A/Melbourne/1/1946(H1N1) Influenza A virus / Strain: A/Melbourne/1/1946(H1N1) |
| Molecular weight | Theoretical: 23.845508 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GLFGAIAGFI EGGWTGMIDG WYGYHHQNEQ GSGYAADQKS TQNAINGITN KVNSVIEKMN TQFTAVGKEF NNLEKRMENL NKKVDDGFL DIWTYNAELL VLLENERTLD FHDSNVKNLY EKVKIQLKNN AKEIGNGCFE FYHKCDNECM ESVRNGTYDY P KYSKEFLV ...String: GLFGAIAGFI EGGWTGMIDG WYGYHHQNEQ GSGYAADQKS TQNAINGITN KVNSVIEKMN TQFTAVGKEF NNLEKRMENL NKKVDDGFL DIWTYNAELL VLLENERTLD FHDSNVKNLY EKVKIQLKNN AKEIGNGCFE FYHKCDNECM ESVRNGTYDY P KYSKEFLV PRGSPGSGYI PEAPRDGQAY VRKDGEWVLL STFLGHHHHH H |
-Macromolecule #3: CR6261 Fab heavy chain
| Macromolecule | Name: CR6261 Fab heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 27.066621 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEWSWVFLFF LSVTTGVHSE VQLVESGAEV KKPGSSVKVS CKASGGPFRS YAISWVRQAP GQGPEWMGGI IPIFGTTKYA PKFQGRVTI TADDFAGTVY MELSSLRSED TAMYYCAKHM GYQVRETMDV WGKGTTVTVS SASTKGPSVF PLAPSSKSTS G GTAALGCL ...String: MEWSWVFLFF LSVTTGVHSE VQLVESGAEV KKPGSSVKVS CKASGGPFRS YAISWVRQAP GQGPEWMGGI IPIFGTTKYA PKFQGRVTI TADDFAGTVY MELSSLRSED TAMYYCAKHM GYQVRETMDV WGKGTTVTVS SASTKGPSVF PLAPSSKSTS G GTAALGCL VKDYFPEPVT VSWNSGALTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYICNVNHK PSNTKVDKRV EP KSCDKHH HHHH |
-Macromolecule #4: CR6261 Fab light chain
| Macromolecule | Name: CR6261 Fab light chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.514395 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEWSWVFLFF LSVTTGVHSQ SVLTQPPSVS AAPGQKVTIS CSGSSSNIGN DYVSWYQQLP GTAPKLLIYD NNKRPSGIPD RFSGSKSGT SATLGITGLQ TGDEANYYCA TWDRRPTAYV VFGGGTKLTV LGAAAGQPKA APSVTLFPPS SEELQANKAT L VCLISDFY ...String: MEWSWVFLFF LSVTTGVHSQ SVLTQPPSVS AAPGQKVTIS CSGSSSNIGN DYVSWYQQLP GTAPKLLIYD NNKRPSGIPD RFSGSKSGT SATLGITGLQ TGDEANYYCA TWDRRPTAYV VFGGGTKLTV LGAAAGQPKA APSVTLFPPS SEELQANKAT L VCLISDFY PGAVTVAWKA DSSPVKAGVE TTTPSKQSNN KYAASSYLSL TPEQWKSHRS YSCQVTHEGS TVEKTVAPTE CS |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 61 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: EMS Lacey Carbon / Material: COPPER / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: LACEY | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 9179 / Average exposure time: 2.4 sec. / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)