[English] 日本語
 Yorodumi
Yorodumi- EMDB-27149: Zebrafish MFSD2A isoform B in inward open ligand-free conformation -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Zebrafish MFSD2A isoform B in inward open ligand-free conformation | ||||||||||||
 Map data Map data | Single-particle cryo-EM map of MFSD2A isoform B from zebrafish with a Fab in the inward open ligand-free conformation at an average resolution of 3.4 A, filtered to local resolution. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | omega-3 fatty acid / lipid transport / membrane protein / Fab | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationSynthesis of PC / lysophospholipid translocation / fatty acid transmembrane transporter activity / lysophospholipid:sodium symporter activity / lysophospholipid transport / lipid transport across blood-brain barrier / transcytosis / establishment of blood-brain barrier / symporter activity / carbohydrate transport ...Synthesis of PC / lysophospholipid translocation / fatty acid transmembrane transporter activity / lysophospholipid:sodium symporter activity / lysophospholipid transport / lipid transport across blood-brain barrier / transcytosis / establishment of blood-brain barrier / symporter activity / carbohydrate transport / fatty acid transport / transmembrane transport / endoplasmic reticulum membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |   | ||||||||||||
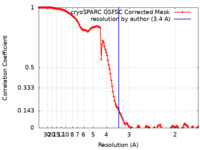
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | ||||||||||||
 Authors Authors | Nguyen C / Lei HT / Lai LTF / Gallentino MJ / Mu X / Matthies D / Gonen T | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Lipid flipping in the omega-3 fatty-acid transporter. Authors: Chi Nguyen / Hsiang-Ting Lei / Louis Tung Faat Lai / Marc J Gallenito / Xuelang Mu / Doreen Matthies / Tamir Gonen /  Abstract: Mfsd2a is the transporter for docosahexaenoic acid (DHA), an omega-3 fatty acid, across the blood brain barrier (BBB). Defects in Mfsd2a are linked to ailments from behavioral and motor dysfunctions ...Mfsd2a is the transporter for docosahexaenoic acid (DHA), an omega-3 fatty acid, across the blood brain barrier (BBB). Defects in Mfsd2a are linked to ailments from behavioral and motor dysfunctions to microcephaly. Mfsd2a transports long-chain unsaturated fatty-acids, including DHA and α-linolenic acid (ALA), that are attached to the zwitterionic lysophosphatidylcholine (LPC) headgroup. Even with the recently determined structures of Mfsd2a, the molecular details of how this transporter performs the energetically unfavorable task of translocating and flipping lysolipids across the lipid bilayer remains unclear. Here, we report five single-particle cryo-EM structures of Danio rerio Mfsd2a (drMfsd2a): in the inward-open conformation in the ligand-free state and displaying lipid-like densities modeled as ALA-LPC at four distinct positions. These Mfsd2a snapshots detail the flipping mechanism for lipid-LPC from outer to inner membrane leaflet and release for membrane integration on the cytoplasmic side. These results also map Mfsd2a mutants that disrupt lipid-LPC transport and are associated with disease. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27149.map.gz emd_27149.map.gz | 5.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27149-v30.xml emd-27149-v30.xml emd-27149.xml emd-27149.xml | 33.8 KB 33.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27149_fsc.xml emd_27149_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_27149.png emd_27149.png | 59.2 KB | ||
| Filedesc metadata |  emd-27149.cif.gz emd-27149.cif.gz | 8.8 KB | ||
| Others |  emd_27149_additional_1.map.gz emd_27149_additional_1.map.gz emd_27149_additional_2.map.gz emd_27149_additional_2.map.gz emd_27149_half_map_1.map.gz emd_27149_half_map_1.map.gz emd_27149_half_map_2.map.gz emd_27149_half_map_2.map.gz | 107.8 MB 204.1 MB 200.8 MB 200.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27149 http://ftp.pdbj.org/pub/emdb/structures/EMD-27149 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27149 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27149 | HTTPS FTP |
-Related structure data
| Related structure data |  8d2tMC  8d2sC  8d2uC  8d2vC  8d2wC  8d2xC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27149.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27149.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single-particle cryo-EM map of MFSD2A isoform B from zebrafish with a Fab in the inward open ligand-free conformation at an average resolution of 3.4 A, filtered to local resolution. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.844 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Single-particle cryo-EM map of MFSD2A isoform B from...
| File | emd_27149_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single-particle cryo-EM map of MFSD2A isoform B from zebrafish with a Fab in the inward open ligand-free conformation at an average resolution of 3.4 A, unsharpened. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Single-particle cryo-EM map of MFSD2A isoform B from...
| File | emd_27149_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single-particle cryo-EM map of MFSD2A isoform B from zebrafish with a Fab in the inward open ligand-free conformation at an average resolution of 3.4 A, sharpened B-114. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Single-particle cryo-EM half map A of MFSD2A isoform...
| File | emd_27149_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single-particle cryo-EM half map A of MFSD2A isoform B from zebrafish with a Fab in the inward open ligand-free conformation. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Single-particle cryo-EM half map B of MFSD2A isoform...
| File | emd_27149_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single-particle cryo-EM half map B of MFSD2A isoform B from zebrafish with a Fab in the inward open ligand-free conformation. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Single-particle cryo-EM map of MFSD2A isoform B from zebrafish wi...
| Entire | Name: Single-particle cryo-EM map of MFSD2A isoform B from zebrafish with a Fab in the inward open ligand-free conformation at an average resolution of 3.4 A, filtered to local resolution |
|---|---|
| Components |
|
-Supramolecule #1: Single-particle cryo-EM map of MFSD2A isoform B from zebrafish wi...
| Supramolecule | Name: Single-particle cryo-EM map of MFSD2A isoform B from zebrafish with a Fab in the inward open ligand-free conformation at an average resolution of 3.4 A, filtered to local resolution type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 56 KDa |
-Macromolecule #1: Sodium-dependent lysophosphatidylcholine symporter 1-B
| Macromolecule | Name: Sodium-dependent lysophosphatidylcholine symporter 1-B type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 56.683965 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHHHH HENLYFQGGS DEVKLAKHET KSRLSVCSKL CYAIGGAPYQ ITGCAIGFFL QIYLLDVALL DPFYASIILF VGRAWDAVT DPTVGFLVSR TPWTRFGRMM PWIVLSTPFA VLCYFLIWYV PSVDQGKVVW YLIFYCCFQT LQTCFHVPYS A LTMFISTE ...String: MHHHHHHHHH HENLYFQGGS DEVKLAKHET KSRLSVCSKL CYAIGGAPYQ ITGCAIGFFL QIYLLDVALL DPFYASIILF VGRAWDAVT DPTVGFLVSR TPWTRFGRMM PWIVLSTPFA VLCYFLIWYV PSVDQGKVVW YLIFYCCFQT LQTCFHVPYS A LTMFISTE QKERDSATAY RMTVEVLGTL IGTAIQGQIV GMANAPCIST EIDLQSTGLE VAPDVQITDP HVSLQDLRNA YM IASGVIC AIYVVCAVVL FLGVKEQKDT CRVRTEPMSF FQGICMVMGH GPYAKLVMGF LFTSLAFMLL EGNFALFCIY NLG FRNDFQ NVLLVIMLSA TLAIPFWQWF LTKFGKKTAV YIGTTSVVPF LISVVLVPSS LAVTYIASFA AGVSVAAAFL LPWS MLPDV VDDFKVQNPE SQGHEAIFYS FYVFFTKFAS GVSLGVSTLS LDFAGYVTRG CTQPGEVKLT LKILVSAAPI VLIII GLLI FISYPINEEK RQGNRKLLNE QREQ UniProtKB: Sodium-dependent lysophosphatidylcholine symporter 1-B |
-Macromolecule #2: FAB light chain
| Macromolecule | Name: FAB light chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 20.674949 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ALDINSPEAE KNAKGARARI TCNAGNQVGS AVAWFNQRPG DPASLLTYWA ATEKGVAGKQ SAQGASTKFS MSSAGPEAPS LSSYWCLLF EKGAFSFGGS KLNPREGAGP QASILPPSAD LNTSGGAAVV CFLPNWYGNI TVQWKTEAPQ SQANMSWPGQ A GANAAYAM ...String: ALDINSPEAE KNAKGARARI TCNAGNQVGS AVAWFNQRPG DPASLLTYWA ATEKGVAGKQ SAQGASTKFS MSSAGPEAPS LSSYWCLLF EKGAFSFGGS KLNPREGAGP QASILPPSAD LNTSGGAAVV CFLPNWYGNI TVQWKTEAPQ SQANMSWPGQ A GANAAYAM AAVLAITKGD YGPGSFTCNA SNRGTGPFAM SLN |
-Macromolecule #3: FAB heavy chain
| Macromolecule | Name: FAB heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 20.568018 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ASKLELSGPA EPRGSKSAQI TCKAKGFPEA RFWVFWLFQR AAALDWPAAN FSGGPVQFES RFQGNASLKG SQAQANAELN IGALGSSTA TYRCGWKLAN GGFFPSWGGA NVNGAAGAKA PAVYPVEISG AGTGSVTLGC LVKGYNAKPN LTWPGASGAL T FPSELNGA ...String: ASKLELSGPA EPRGSKSAQI TCKAKGFPEA RFWVFWLFQR AAALDWPAAN FSGGPVQFES RFQGNASLKG SQAQANAELN IGALGSSTA TYRCGWKLAN GGFFPSWGGA NVNGAAGAKA PAVYPVEISG AGTGSVTLGC LVKGYNAKPN LTWPGASGAL T FPSELNGA LWNLASAVTG SGFPSATCAV GFGAATDVDK KVAAA |
-Macromolecule #4: DODECYL-BETA-D-MALTOSIDE
| Macromolecule | Name: DODECYL-BETA-D-MALTOSIDE / type: ligand / ID: 4 / Number of copies: 10 / Formula: LMT |
|---|---|
| Molecular weight | Theoretical: 510.615 Da |
| Chemical component information |  ChemComp-LMT: |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 4 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: 50 mM HEPES (pH 8), 150 mM NaCl and 0.02% DDM | ||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: AIR | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 86 % / Chamber temperature: 277 K / Instrument: LEICA EM GP Details: 400-mesh 1.2/1.3 Cu grids (Quantifoil) were made hydrophilic by glow discharging for two times 60 seconds with a current of 15 mA in a PELCO easiGlow system. The cryo grids were produced ...Details: 400-mesh 1.2/1.3 Cu grids (Quantifoil) were made hydrophilic by glow discharging for two times 60 seconds with a current of 15 mA in a PELCO easiGlow system. The cryo grids were produced using a Leica EM GP (Leica). The chamber was kept at 4 C and 95% humidity (86-91% measured). 3 microliter sample at 3 mg/ml was applied to a glow-discharged holey grid, blotted for 6 s, and plunge frozen into liquid ethane and stored in liquid nitrogen.. | ||||||||
| Details | Purified 15A9 FAB fragment was incubated overnight at 4 C with MFSD2A at a 5:1 molar ratio. DrMFSD2A-15A9 complex was injected into the size exclusion column Superdex200 to separate the unbound FAB. The size exclusion column step was also used to exchange the buffer of the MFSD2A-FAB complex into 50 mM HEPES, 150 mM NaCl and 0.02% DDM. SDS-PAGE was used to pool peak fractions containing both MFSD2A and FAB, indicating complex formation. The pooled fractions containing MFSD2A-FAB were concentrated to 3 mg/ml using Amicon spin concentrator with a 30 kDa cutoff. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Details | Cryo-EM grids were loaded into a 300 keV FEI Titan Krios cryo electron microscope (ThermoFisher Scientific, formerly FEI) at HHMI Janelia Reasearch Campus, Janelia Krios 1, equipped with a Cs corrector, and Gatan energy filter and K3 camera (Gatan Inc.). Movies of 50 frames with 1 e-/A2 per frame (50 e-/A2 total dose) were automatically recorded at a nominal magnification of 81,000x, corresponding to a physical pixel size of 0.844 A/px (superresolution pixel size 0.422 A/px) in CDS mode at a dose rate of 9.5 e-/px/s (~7.5 e-/px/s on the camera through the sample) and a defocus range of -0.5 to -1.8 micrometer using SerialEM. In total, 2,653 and 8,443 movies were collected in two separate imaging sessions, with the first dataset on a 400-mesh copper grid and the second on a 300-mesh UltrAuFoil gold grid. |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 2 / Number real images: 11096 / Average exposure time: 3.75 sec. / Average electron dose: 50.0 e/Å2 Details: Movies of 50 frames with 1 e-/A2 per frame (50 e-/A2 total dose) were automatically recorded at a nominal magnification of 81,000x, corresponding to a physical pixel size of 0.844 A/px ...Details: Movies of 50 frames with 1 e-/A2 per frame (50 e-/A2 total dose) were automatically recorded at a nominal magnification of 81,000x, corresponding to a physical pixel size of 0.844 A/px (superresolution pixel size 0.422 A/px) in CDS mode at a dose rate of 9.5 e-/px/s (~7.5 e-/px/s on the camera through the sample) and a defocus range of -0.5 to -1.8 mircometer using SerialEM. In total, 2,653 and 8,443 movies were collected in two separate imaging sessions, with the first dataset on a 400-mesh copper grid and the second on a 300-mesh UltrAuFoil gold grid. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-8d2t: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)