+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of TTMV-LY1 anellovirus virus-like particle | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | anellovirus / VIRUS LIKE PARTICLE | |||||||||
| Function / homology | TT viral protein of unknown function / TT viral orf 1 / T=1 icosahedral viral capsid / Capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |  Anelloviridae (virus) / Anelloviridae (virus) /  TTV-like mini virus TTV-like mini virus | |||||||||
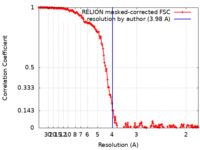
| Method | single particle reconstruction / cryo EM / Resolution: 3.98 Å | |||||||||
 Authors Authors | Liou SH / Delagrave S / Swanson K | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structure of anellovirus-like particles reveal a mechanism for immune evasion. Authors: Shu-Hao Liou / Rajendra Boggavarapu / Noah R Cohen / Yue Zhang / Ishwari Sharma / Lynn Zeheb / Nidhi Mukund Acharekar / Hillary D Rodgers / Saadman Islam / Jared Pitts / Cesar Arze / Harish ...Authors: Shu-Hao Liou / Rajendra Boggavarapu / Noah R Cohen / Yue Zhang / Ishwari Sharma / Lynn Zeheb / Nidhi Mukund Acharekar / Hillary D Rodgers / Saadman Islam / Jared Pitts / Cesar Arze / Harish Swaminathan / Nathan Yozwiak / Tuyen Ong / Roger J Hajjar / Yong Chang / Kurt A Swanson / Simon Delagrave /  Abstract: Anelloviruses are nonpathogenic viruses that comprise a major portion of the human virome. Despite being ubiquitous in the human population, anelloviruses (ANVs) remain poorly understood. Basic ...Anelloviruses are nonpathogenic viruses that comprise a major portion of the human virome. Despite being ubiquitous in the human population, anelloviruses (ANVs) remain poorly understood. Basic features of the virus, such as the identity of its capsid protein and the structure of the viral particle, have been unclear until now. Here, we use cryogenic electron microscopy to describe the first structure of an ANV-like particle. The particle, formed by 60 jelly roll domain-containing ANV capsid proteins, forms an icosahedral particle core from which spike domains extend to form a salient part of the particle surface. The spike domains come together around the 5-fold symmetry axis to form crown-like features. The base of the spike domain, the P1 subdomain, shares some sequence conservation between ANV strains while a hypervariable region, forming the P2 subdomain, is at the spike domain apex. We propose that this structure renders the particle less susceptible to antibody neutralization by hiding vulnerable conserved domains while exposing highly diverse epitopes as immunological decoys, thereby contributing to the immune evasion properties of anelloviruses. These results shed light on the structure of anelloviruses and provide a framework to understand their interactions with the immune system. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27077.map.gz emd_27077.map.gz | 237.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27077-v30.xml emd-27077-v30.xml emd-27077.xml emd-27077.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27077_fsc.xml emd_27077_fsc.xml | 15.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_27077.png emd_27077.png | 37.7 KB | ||
| Filedesc metadata |  emd-27077.cif.gz emd-27077.cif.gz | 5.5 KB | ||
| Others |  emd_27077_half_map_1.map.gz emd_27077_half_map_1.map.gz emd_27077_half_map_2.map.gz emd_27077_half_map_2.map.gz | 258.8 MB 258.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27077 http://ftp.pdbj.org/pub/emdb/structures/EMD-27077 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27077 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27077 | HTTPS FTP |
-Related structure data
| Related structure data |  8cygMC  8v7xC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27077.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27077.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.923 Å | ||||||||||||||||||||||||||||||||||||
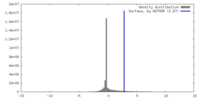
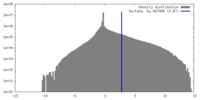
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : TTV-like mini virus
| Entire | Name:  TTV-like mini virus TTV-like mini virus |
|---|---|
| Components |
|
-Supramolecule #1: TTV-like mini virus
| Supramolecule | Name: TTV-like mini virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 93678 / Sci species name: TTV-like mini virus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: No / Virus empty: Yes |
|---|
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Details: TTV-like mini virus isolate TTMV_LY1 / Number of copies: 60 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Anelloviridae (virus) Anelloviridae (virus) |
| Molecular weight | Theoretical: 79.869227 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPWWYRRRSY NPWRRRNWFR RPRKTIYRRY RRRRRWVRRK PFYKRKIKRL NIVEWQPKSI RKCRIKGMLC LFQTTEDRLS YNFDMYEES IIPEKLPGGG GFSIKNISLY ALYQEHIHAH NIFTHTNTDR PLARYTGCSL KFYQSKDIDY VVTYSTSLPL R SSMGMYNS ...String: MPWWYRRRSY NPWRRRNWFR RPRKTIYRRY RRRRRWVRRK PFYKRKIKRL NIVEWQPKSI RKCRIKGMLC LFQTTEDRLS YNFDMYEES IIPEKLPGGG GFSIKNISLY ALYQEHIHAH NIFTHTNTDR PLARYTGCSL KFYQSKDIDY VVTYSTSLPL R SSMGMYNS MQPSIHLMQQ NKLIVPSKQT QKRRKPYIKK HISPPTQMKS QWYFQHNIAN IPLLMIRTTA LTLDNYYIGS RQ LSTNVTI HTLNTTYIQN RDWGDRNKTY YCQTLGTQRY FLYGTHSTAQ NINDIKLQEL IPLTNTQDYV QGFDWTEKDK HNI TTYKEF LTKGAGNPFH AEWITAQNPV IHTANSPTQI EQIYTASTTT FQNKKLTDLP TPGYIFITPT VSLRYNPYKD LAER NKCYF VRSKINAHGW DPEQHQELIN SDLPQWLLLF GYPDYIKRTQ NFALVDTNYI LVDHCPYTNP EKTPFIPLST SFIEG RSPY SPSDTHEPDE EDQNRWYPCY QYQQESINSI CLSGPGTPKI PKGITAEAKV KYSFNFKWGG DLPPMSTITN PTDQPT YVV PNNFNETTSL QNPTTRPEHF LYSFDERRGQ LTEKATKRLL KDWETKETSL LSTEYRFAEP TQTQAPQEDP SSEEEEE SN LFERLLRQRT KQLQLKRRII QTLKDLQKLE UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 19.59 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
 Movie
Movie Controller
Controller






 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)