[English] 日本語
 Yorodumi
Yorodumi- EMDB-26496: Cryo-EM structure of BG24 Fabs with an inferred germline CDRL1 an... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
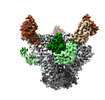
| Title | Cryo-EM structure of BG24 Fabs with an inferred germline CDRL1 and 10-1074 Fabs in complex with HIV-1 Env 6405-SOSIP.664 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV-1 Env / neutralizing antibody / CD4 binding site antibody / VIRAL PROTEIN / Germline / precursor / immunogen / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / virus-mediated perturbation of host defense response / fusion of virus membrane with host endosome membrane / viral envelope ...positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / virus-mediated perturbation of host defense response / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / apoptotic process / host cell plasma membrane / structural molecule activity / virion membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Dam KA / Bjorkman PJ | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: HIV-1 CD4-binding site germline antibody-Env structures inform vaccine design. Authors: Kim-Marie A Dam / Christopher O Barnes / Harry B Gristick / Till Schoofs / Priyanthi N P Gnanapragasam / Michel C Nussenzweig / Pamela J Bjorkman /    Abstract: BG24, a VRC01-class broadly neutralizing antibody (bNAb) against HIV-1 Env with relatively few somatic hypermutations (SHMs), represents a promising target for vaccine strategies to elicit CD4- ...BG24, a VRC01-class broadly neutralizing antibody (bNAb) against HIV-1 Env with relatively few somatic hypermutations (SHMs), represents a promising target for vaccine strategies to elicit CD4-binding site (CD4bs) bNAbs. To understand how SHMs correlate with BG24 neutralization of HIV-1, we report 4.1 Å and 3.4 Å single-particle cryo-EM structures of two inferred germline (iGL) BG24 precursors complexed with engineered Env-based immunogens lacking CD4bs N-glycans. Structures reveal critical Env contacts by BG24 and identify antibody light chain structural features that impede Env recognition. In addition, biochemical data and cryo-EM structures of BG24 variants bound to Envs with CD4bs glycans present provide insights into N-glycan accommodation, including structural modes of light chain adaptations in the presence of the N276 glycan. Together, these findings reveal Env regions critical for germline antibody recognition and potential sites to alter in immunogen design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26496.map.gz emd_26496.map.gz | 17.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26496-v30.xml emd-26496-v30.xml emd-26496.xml emd-26496.xml | 22.3 KB 22.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26496.png emd_26496.png | 106.3 KB | ||
| Filedesc metadata |  emd-26496.cif.gz emd-26496.cif.gz | 6.7 KB | ||
| Others |  emd_26496_additional_1.map.gz emd_26496_additional_1.map.gz emd_26496_half_map_1.map.gz emd_26496_half_map_1.map.gz emd_26496_half_map_2.map.gz emd_26496_half_map_2.map.gz | 167.7 MB 140.7 MB 140.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26496 http://ftp.pdbj.org/pub/emdb/structures/EMD-26496 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26496 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26496 | HTTPS FTP |
-Validation report
| Summary document |  emd_26496_validation.pdf.gz emd_26496_validation.pdf.gz | 793.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26496_full_validation.pdf.gz emd_26496_full_validation.pdf.gz | 793.4 KB | Display | |
| Data in XML |  emd_26496_validation.xml.gz emd_26496_validation.xml.gz | 15 KB | Display | |
| Data in CIF |  emd_26496_validation.cif.gz emd_26496_validation.cif.gz | 17.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26496 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26496 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26496 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26496 | HTTPS FTP |
-Related structure data
| Related structure data |  7ugqMC  7ugmC  7ugnC  7ugoC  7ugpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26496.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26496.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.869 Å | ||||||||||||||||||||||||||||||||||||
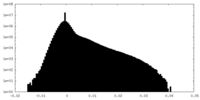
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Local refinement map used to build the BG24...
| File | emd_26496_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local refinement map used to build the BG24 with an inferred germline CDRL1 Fab CDRL1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
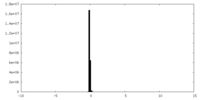
| Density Histograms |
-Half map: #2
| File | emd_26496_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
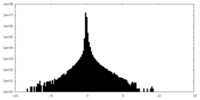
| Density Histograms |
-Half map: #1
| File | emd_26496_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
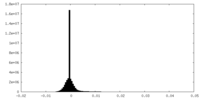
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM map of BG24 Fabs with an inferred germline CDRL1 and 10-1...
| Entire | Name: Cryo-EM map of BG24 Fabs with an inferred germline CDRL1 and 10-1074 Fabs in complex with HIV-1 Env 6405-SOSIP.664 |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM map of BG24 Fabs with an inferred germline CDRL1 and 10-1...
| Supramolecule | Name: Cryo-EM map of BG24 Fabs with an inferred germline CDRL1 and 10-1074 Fabs in complex with HIV-1 Env 6405-SOSIP.664 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: Envelope glycoprotein gp120
| Macromolecule | Name: Envelope glycoprotein gp120 / type: protein_or_peptide / ID: 1 / Details: Env mimic 6405-SOSIP.664 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 50.007793 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: NLWVTVYYGV PVWKEAKTTL FCASDAKSYA HNIWATHACV PTDPNPQEVE LVNVTENFNM WKNDMVDQMH EDIISLWDES LKPCVKLTP LCVTLQCSEM KNGTDRMNCS FNATTVVNDK QKKVHALFYR LDIEPIGNNS TEYMLINCNT SAIAQSCPKI T FEPIPIHY ...String: NLWVTVYYGV PVWKEAKTTL FCASDAKSYA HNIWATHACV PTDPNPQEVE LVNVTENFNM WKNDMVDQMH EDIISLWDES LKPCVKLTP LCVTLQCSEM KNGTDRMNCS FNATTVVNDK QKKVHALFYR LDIEPIGNNS TEYMLINCNT SAIAQSCPKI T FEPIPIHY CAPAGFALLK CRDKQFNGTG PCTNVSSVQC THGIKPVVST QLLLNGSLAE EDIIIRSENL TDNTKTIIVQ LN KSIAINC TRPYNNTRQR IHITNDARVI VGDIRQAHCN VSRVDWNNMT QWAATKLGSL YNRSTIIFNH ASGGDPEITT HSF TCGGEF FYCNTSGLFN STWDIMNTSS PNNTDPIITL QCRIKQIINM WQGVGRAIYA PPIAGQILCS SNITGLLLTR DGGA DNSSH NETFRPGGGN MKDNWRSELY KYKVVKIEPL GVAPTRCKRR VV UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: Envelope glycoprotein gp41
| Macromolecule | Name: Envelope glycoprotein gp41 / type: protein_or_peptide / ID: 2 / Details: Env mimic 6405-SOSIP.664 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 14.676723 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VFLGFLGAAG STMGAASMTL TVQARNLLSL LKLTVWGIKQ LQARVLAVER YLRDQQLLGI WGCSGKLICC TNVPWNSSWS NRNLSEIWD NMTWLQWDKE ISNYTQIIYG LLEESQNQQE KNEQDLLALD |
-Macromolecule #3: BG24 with an inferred germline CDRL1 Fab heavy chain
| Macromolecule | Name: BG24 with an inferred germline CDRL1 Fab heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.758364 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSRAE VKKPGASVKV SCEASGYNFV DHYIHWVRQA PGQAPQWVGW MNPRGGGVAY SQRFQGRVTM TRDTSIDTAY MQLNRLTSG DTAVYYCATQ VKLDSSAGYP FDIWGQGTMV TVSSAS |
-Macromolecule #4: BG24 with an inferred germline CDRL1 Fab light chain
| Macromolecule | Name: BG24 with an inferred germline CDRL1 Fab light chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.177191 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SALTQPRSVS GSPGQSVNIS CTGTSSDVGG YNYVSWYQQH PGRAPKLIIY EVNRRPSGVS DRFSGSKSGN TASLTISGLR TEDEADYFC SAFEYFGGGT KLTVLS |
-Macromolecule #5: 10-1074 Fab heavy chain
| Macromolecule | Name: 10-1074 Fab heavy chain / type: protein_or_peptide / ID: 5 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.791433 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLQESGPG LVKPSETLSV TCSVSGDSMN NYYWTWIRQS PGKGLEWIGY ISDRESATYN PSLNSRVVIS RDTSKNQLSL KLNSVTPAD TAVYYCATAR RGQRIYGVVS FGEFFYYYSM DVWGKGTTVT VSSA |
-Macromolecule #6: 10-1074 Fab light chain
| Macromolecule | Name: 10-1074 Fab light chain / type: protein_or_peptide / ID: 6 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.701023 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VRPLSVALGE TARISCGRQA LGSRAVQWYQ HRPGQAPILL IYNNQDRPSG IPERFSGTPD INFGTRATLT ISGVEAGDEA DYYCHMWDS RSGFSWSFGG ATRLTVLG |
-Macromolecule #12: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 12 / Number of copies: 24 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: Details: Closed conformation Env trimer (gp120/gp41 subunits) low-pass filtered to 80 angstrom |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 170897 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)