[English] 日本語
 Yorodumi
Yorodumi- EMDB-26183: Cryo-EM Structure of insulin receptor-related receptor (IRR) in a... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
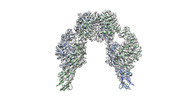
| Title | Cryo-EM Structure of insulin receptor-related receptor (IRR) in apo-state captured at pH 7. The 3D refinement was applied with C2 symmetry | |||||||||
 Map data Map data | Cryo-EM Structure of insulin receptor-related receptor (IRR) in apo-state captured at pH 7. The 3D refinement was applied with C2 symmetry. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Receptor tyrosine kinase / insulin receptor family / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to alkaline pH / male sex determination / insulin receptor complex / insulin receptor activity / insulin receptor substrate binding / phosphatidylinositol 3-kinase binding / transmembrane receptor protein tyrosine kinase activity / cell surface receptor protein tyrosine kinase signaling pathway / receptor protein-tyrosine kinase / insulin receptor signaling pathway ...cellular response to alkaline pH / male sex determination / insulin receptor complex / insulin receptor activity / insulin receptor substrate binding / phosphatidylinositol 3-kinase binding / transmembrane receptor protein tyrosine kinase activity / cell surface receptor protein tyrosine kinase signaling pathway / receptor protein-tyrosine kinase / insulin receptor signaling pathway / protein autophosphorylation / actin cytoskeleton organization / receptor complex / axon / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Wang LW / Hall C / Li J / Choi E / Bai XC | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor. Authors: Liwei Wang / Catherine Hall / Jie Li / Eunhee Choi / Xiao-Chen Bai /  Abstract: The insulin receptor (IR) family is a subfamily of receptor tyrosine kinases that controls metabolic homeostasis and cell growth. Distinct from IR and insulin-like growth factor 1 receptor, whose ...The insulin receptor (IR) family is a subfamily of receptor tyrosine kinases that controls metabolic homeostasis and cell growth. Distinct from IR and insulin-like growth factor 1 receptor, whose activation requires ligand binding, insulin receptor-related receptor (IRR)-the third member of the IR family-is activated by alkaline pH. However, the molecular mechanism underlying alkaline pH-induced IRR activation remains unclear. Here, we present cryo-EM structures of human IRR in both neutral pH inactive and alkaline pH active states. Combined with mutagenesis and cellular assays, we show that, upon pH increase, electrostatic repulsion of the pH-sensitive motifs of IRR disrupts its autoinhibited state and promotes a scissor-like rotation between two protomers, leading to a T-shaped active conformation. Together, our study reveals an unprecedented alkaline pH-dependent activation mechanism of IRR, opening up opportunities to understand the structure-function relationship of this important receptor. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26183.map.gz emd_26183.map.gz | 49.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26183-v30.xml emd-26183-v30.xml emd-26183.xml emd-26183.xml | 13.1 KB 13.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26183.png emd_26183.png | 46.8 KB | ||
| Filedesc metadata |  emd-26183.cif.gz emd-26183.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26183 http://ftp.pdbj.org/pub/emdb/structures/EMD-26183 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26183 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26183 | HTTPS FTP |
-Validation report
| Summary document |  emd_26183_validation.pdf.gz emd_26183_validation.pdf.gz | 593.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26183_full_validation.pdf.gz emd_26183_full_validation.pdf.gz | 593.2 KB | Display | |
| Data in XML |  emd_26183_validation.xml.gz emd_26183_validation.xml.gz | 6 KB | Display | |
| Data in CIF |  emd_26183_validation.cif.gz emd_26183_validation.cif.gz | 6.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26183 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26183 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26183 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26183 | HTTPS FTP |
-Related structure data
| Related structure data |  7tykMC  7tyjC  7tymC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26183.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26183.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM Structure of insulin receptor-related receptor (IRR) in apo-state captured at pH 7. The 3D refinement was applied with C2 symmetry. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
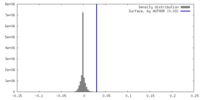
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Insulin receptor-related receptor (IRR) in apo-state captured at pH 7
| Entire | Name: Insulin receptor-related receptor (IRR) in apo-state captured at pH 7 |
|---|---|
| Components |
|
-Supramolecule #1: Insulin receptor-related receptor (IRR) in apo-state captured at pH 7
| Supramolecule | Name: Insulin receptor-related receptor (IRR) in apo-state captured at pH 7 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 140 KDa |
-Macromolecule #1: Insulin receptor-related protein
| Macromolecule | Name: Insulin receptor-related protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: receptor protein-tyrosine kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 143.879547 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAVPSLWPWG ACLPVIFLSL GFGLDTVEVC PSLDIRSEVA ELRQLENCSV VEGHLQILLM FTATGEDFRG LSFPRLTQVT DYLLLFRVY GLESLRDLFP NLAVIRGTRL FLGYALVIFE MPHLRDVALP ALGAVLRGAV RVEKNQELCH LSTIDWGLLQ P APGANHIV ...String: MAVPSLWPWG ACLPVIFLSL GFGLDTVEVC PSLDIRSEVA ELRQLENCSV VEGHLQILLM FTATGEDFRG LSFPRLTQVT DYLLLFRVY GLESLRDLFP NLAVIRGTRL FLGYALVIFE MPHLRDVALP ALGAVLRGAV RVEKNQELCH LSTIDWGLLQ P APGANHIV GNKLGEECAD VCPGVLGAAG EPCAKTTFSG HTDYRCWTSS HCQRVCPCPH GMACTARGEC CHTECLGGCS QP EDPRACV ACRHLYFQGA CLWACPPGTY QYESWRCVTA ERCASLHSVP GRASTFGIHQ GSCLAQCPSG FTRNSSSIFC HKC EGLCPK ECKVGTKTID SIQAAQDLVG CTHVEGSLIL NLRQGYNLEP QLQHSLGLVE TITGFLKIKH SFALVSLGFF KNLK LIRGD AMVDGNYTLY VLDNQNLQQL GSWVAAGLTI PVGKIYFAFN PRLCLEHIYR LEEVTGTRGR QNKAEINPRT NGDRA ACQT RTLRFVSNVT EADRILLRWE RYEPLEARDL LSFIVYYKES PFQNATEHVG PDACGTQSWN LLDVELPLSR TQEPGV TLA SLKPWTQYAV FVRAITLTTE EDSPHQGAQS PIVYLRTLPA APTVPQDVIS TSNSSSHLLV RWKPPTQRNG NLTYYLV LW QRLAEDGDLY LNDYCHRGLR LPTSNNDPRF DGEDGDPEAE MESDCCPCQH PPPGQVLPPL EAQEASFQKK FENFLHNA I TIPISPWKVT SINKSPQRDS GRHRRAAGPL RLGGNSSDFE IQEDKVPRER AVLSGLRHFT EYRIDIHACN HAAHTVGCS AATFVFARTM PHREADGIPG KVAWEASSKN SVLLRWLEPP DPNGLILKYE IKYRRLGEEA TVLCVSRLRY AKFGGVHLAL LPPGNYSAR VRATSLAGNG SWTDSVAFYI LGPEEEDAGG LHVLLTATPV GLTLLIVLAA LGFFYGKKRN RTLYASVNPE Y FSASDMYV PDEWEVPREQ ISIIRELGQG SFGMVYEGLA RGLEAGEEST PVALKTVNEL ASPRECIEFL KEASVMKAFK CH HVVRLLG VVSQGQPTLV IMELMTRGDL KSHLRSLRPE AENNPGLPQP ALGEMIQMAG EIADGMAYLA ANKFVHRDLA ARN CMVSQD FTVKIGDFGM TRDVYETDYY RKGGKGLLPV RWMAPESLKD GIFTTHSDVW SFGVVLWEIV TLAEQPYQGL SNEQ VLKFV MDGGVLEELE GCPLQLQELM SRCWQPNPRL RPSFTHILDS IQEELRPSFR LLSFYYSPEC RGARGSLPTT DAEPD SSPT PRDCSPQNGG PGH UniProtKB: Insulin receptor-related protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.6 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7tyk: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)