[English] 日本語
 Yorodumi
Yorodumi- EMDB-24792: M. tuberculosis ribosomal RNA methyltransferase TlyA bound to M. ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | M. tuberculosis ribosomal RNA methyltransferase TlyA bound to M. smegmatis 50S ribosomal subunit | |||||||||
 Map data Map data | Mtb TlyA bound to Msg 50S ribosomal subunit. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ribosome / Methyltransferase / rRNA modification / 50S / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology information23S rRNA (cytidine1920-2'-O)-methyltransferase / 16S rRNA (cytidine1409-2'-O)-methyltransferase / methyltransferase activity / methylation / cytosolic large ribosomal subunit / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation ...23S rRNA (cytidine1920-2'-O)-methyltransferase / 16S rRNA (cytidine1409-2'-O)-methyltransferase / methyltransferase activity / methylation / cytosolic large ribosomal subunit / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / mRNA binding / RNA binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Mycolicibacterium smegmatis (bacteria) / Mycolicibacterium smegmatis (bacteria) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.05 Å | |||||||||
 Authors Authors | Laughlin ZT / Dunham CM | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: 50S subunit recognition and modification by the ribosomal RNA methyltransferase TlyA. Authors: Zane T Laughlin / Suparno Nandi / Debayan Dey / Natalia Zelinskaya / Marta A Witek / Pooja Srinivas / Ha An Nguyen / Emily G Kuiper / Lindsay R Comstock / Christine M Dunham / Graeme L Conn /  Abstract: Changes in bacterial ribosomal RNA (rRNA) methylation status can alter the activity of diverse groups of ribosome-targeting antibiotics. These modifications are typically incorporated by a single ...Changes in bacterial ribosomal RNA (rRNA) methylation status can alter the activity of diverse groups of ribosome-targeting antibiotics. These modifications are typically incorporated by a single methyltransferase that acts on one nucleotide target and rRNA methylation directly prevents drug binding, thereby conferring drug resistance. Loss of intrinsic methylation can also result in antibiotic resistance. For example, Mycobacterium tuberculosis becomes sensitized to tuberactinomycin antibiotics, such as capreomycin and viomycin, due to the action of the intrinsic methyltransferase TlyA. TlyA is unique among antibiotic resistance-associated methyltransferases as it has dual 16S and 23S rRNA substrate specificity and can incorporate cytidine-2′-O-methylations within two structurally distinct contexts. Here, we report the structure of a mycobacterial 50S subunit-TlyA complex trapped in a postcatalytic state with a S-adenosyl-L-methionine analog using single-particle cryogenic electron microscopy. Together with complementary functional analyses, this structure reveals critical roles in 23S rRNA substrate recognition for conserved residues across an interaction surface that spans both TlyA domains. These interactions position the TlyA active site over the target nucleotide C2144, which is flipped from 23S Helix 69 in a process stabilized by stacking of TlyA residue Phe157 on the adjacent A2143. Base flipping may thus be a common strategy among rRNA methyltransferase enzymes, even in cases where the target site is accessible without such structural reorganization. Finally, functional studies with 30S subunit suggest that the same TlyA interaction surface is employed to recognize this second substrate, but with distinct dependencies on essential conserved residues. #1:  Journal: Biorxiv / Year: 2022 Journal: Biorxiv / Year: 2022Title: 50S subunit recognition and modification by the Mycobacterium tuberculosis ribosomal RNA methyltransferase TlyA Authors: Laughlin ZT / Nandi S / Dey D / Zelinskaya N / Witek MA / Srinivas P / Nguyen HA / Kuiper EG / Comstock LR / Dunham CM / Conn GL | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24792.map.gz emd_24792.map.gz | 78.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24792-v30.xml emd-24792-v30.xml emd-24792.xml emd-24792.xml | 62.4 KB 62.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24792_fsc.xml emd_24792_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_24792.png emd_24792.png | 82.3 KB | ||
| Filedesc metadata |  emd-24792.cif.gz emd-24792.cif.gz | 12.7 KB | ||
| Others |  emd_24792_additional_1.map.gz emd_24792_additional_1.map.gz emd_24792_additional_2.map.gz emd_24792_additional_2.map.gz | 77.3 MB 78.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24792 http://ftp.pdbj.org/pub/emdb/structures/EMD-24792 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24792 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24792 | HTTPS FTP |
-Related structure data
| Related structure data |  7s0sMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24792.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24792.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mtb TlyA bound to Msg 50S ribosomal subunit. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.069 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Mtb TlyA bound to H69 and proximal rRNA...
| File | emd_24792_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mtb TlyA bound to H69 and proximal rRNA without the rest of Msg 50S. Created from multi body refinement and used in model building and refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
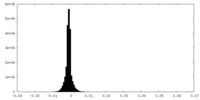
| Density Histograms |
-Additional map: Msg 50S alone without H69 or Mtb TlyA....
| File | emd_24792_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Msg 50S alone without H69 or Mtb TlyA. Created from multi body refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
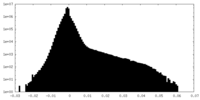
| Density Histograms |
- Sample components
Sample components
+Entire : Mtb TlyA bound to Msg 50S ribosomal subunit modified at C2144 wit...
+Supramolecule #1: Mtb TlyA bound to Msg 50S ribosomal subunit modified at C2144 wit...
+Supramolecule #2: 50S
+Supramolecule #3: TlyA
+Macromolecule #1: ribosomal protein bL37
+Macromolecule #2: 16S/23S rRNA (Cytidine-2'-O)-methyltransferase TlyA
+Macromolecule #4: 50S ribosomal protein L2
+Macromolecule #5: 50S ribosomal protein L3
+Macromolecule #6: 50S ribosomal protein L4
+Macromolecule #7: 50S ribosomal protein L5
+Macromolecule #8: 50S ribosomal protein L6
+Macromolecule #9: 50S ribosomal protein L9
+Macromolecule #10: 50S ribosomal protein L10
+Macromolecule #11: 50S ribosomal protein L11
+Macromolecule #12: 50S ribosomal protein L13
+Macromolecule #13: 50S ribosomal protein L14
+Macromolecule #14: 50S ribosomal protein L15
+Macromolecule #15: 50S ribosomal protein L16
+Macromolecule #16: 50S ribosomal protein L17
+Macromolecule #17: 50S ribosomal protein L18
+Macromolecule #18: 50S ribosomal protein L19
+Macromolecule #19: 50S ribosomal protein L20
+Macromolecule #20: 50S ribosomal protein L21
+Macromolecule #21: 50S ribosomal protein L22
+Macromolecule #22: 50S ribosomal protein L23
+Macromolecule #23: 50S ribosomal protein L24
+Macromolecule #24: 50S ribosomal protein L25
+Macromolecule #25: 50S ribosomal protein L27
+Macromolecule #26: 50S ribosomal protein L28
+Macromolecule #27: 50S ribosomal protein L29
+Macromolecule #28: 50S ribosomal protein L30
+Macromolecule #29: 50S ribosomal protein L32
+Macromolecule #30: 50S ribosomal protein L33
+Macromolecule #31: 50S ribosomal protein L34
+Macromolecule #32: 50S ribosomal protein L35
+Macromolecule #33: 50S ribosomal protein L36
+Macromolecule #34: 50S ribosomal protein L31
+Macromolecule #3: 23S rRNA
+Macromolecule #35: 5S rRNA
+Macromolecule #36: MAGNESIUM ION
+Macromolecule #37: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 Component:
Details: Both components dialyzed into this buffer | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 298 K / Instrument: GATAN CRYOPLUNGE 3 / Details: 3.0 or 3.3 second blot time allowed for sample. | ||||||||||||
| Details | 0.5 micromolar 50S, 5 micromolar TlyA, 10 micromolar NM6 |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 2 / Number real images: 3364 / Average exposure time: 2.0 sec. / Average electron dose: 50.79 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 81000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Output model |  PDB-7s0s: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)