[English] 日本語
 Yorodumi
Yorodumi- EMDB-24201: Late pre-fusion state of EEEV (pH 5.5) with localized reconstruction -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Late pre-fusion state of EEEV (pH 5.5) with localized reconstruction | ||||||||||||||||||
 Map data Map data | Localized reconstruction of EEEV at acidic pH (late prefusion) | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | EEEV / pre-fusion / localized reconstruction / VIRUS | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationtogavirin / T=4 icosahedral viral capsid / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / symbiont-mediated suppression of host gene expression / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / symbiont entry into host cell / virion attachment to host cell / host cell nucleus ...togavirin / T=4 icosahedral viral capsid / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / symbiont-mediated suppression of host gene expression / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / symbiont entry into host cell / virion attachment to host cell / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / RNA binding / membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  Eastern equine encephalitis virus (strain Florida 91-469) / Eastern equine encephalitis virus (strain Florida 91-469) /   Eastern equine encephalitis virus Eastern equine encephalitis virus | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 14.1 Å | ||||||||||||||||||
 Authors Authors | Chen C-L / Kuhn RJ / Klose T | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Cryo-EM structures of alphavirus conformational intermediates in low pH-triggered prefusion states. Authors: Chun-Liang Chen / Thomas Klose / Chengqun Sun / Arthur S Kim / Geeta Buda / Michael G Rossmann / Michael S Diamond / William B Klimstra / Richard J Kuhn /  Abstract: Alphaviruses can cause severe human arthritis and encephalitis. During virus infection, structural changes of viral glycoproteins in the acidified endosome trigger virus-host membrane fusion for ...Alphaviruses can cause severe human arthritis and encephalitis. During virus infection, structural changes of viral glycoproteins in the acidified endosome trigger virus-host membrane fusion for delivery of the capsid core and RNA genome into the cytosol to initiate virus translation and replication. However, mechanisms by which E1 and E2 glycoproteins rearrange in this process remain unknown. Here, we investigate prefusion cryoelectron microscopy (cryo-EM) structures of eastern equine encephalitis virus (EEEV) under acidic conditions. With models fitted into the low-pH cryo-EM maps, we suggest that E2 dissociates from E1, accompanied by a rotation (∼60°) of the E2-B domain (E2-B) to expose E1 fusion loops. Cryo-EM reconstructions of EEEV bound to a protective antibody at acidic and neutral pH suggest that stabilization of E2-B prevents dissociation of E2 from E1. These findings reveal conformational changes of the glycoprotein spikes in the acidified host endosome. Stabilization of E2-B may provide a strategy for antiviral agent development. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24201.map.gz emd_24201.map.gz | 10.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24201-v30.xml emd-24201-v30.xml emd-24201.xml emd-24201.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24201_fsc.xml emd_24201_fsc.xml | 5.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_24201.png emd_24201.png | 77.6 KB | ||
| Filedesc metadata |  emd-24201.cif.gz emd-24201.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24201 http://ftp.pdbj.org/pub/emdb/structures/EMD-24201 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24201 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24201 | HTTPS FTP |
-Related structure data
| Related structure data |  7n69MC  7n6aC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24201.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24201.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Localized reconstruction of EEEV at acidic pH (late prefusion) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
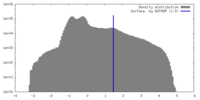
| Voxel size | X=Y=Z: 3.496 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Eastern equine encephalitis virus
| Entire | Name:   Eastern equine encephalitis virus Eastern equine encephalitis virus |
|---|---|
| Components |
|
-Supramolecule #1: Eastern equine encephalitis virus
| Supramolecule | Name: Eastern equine encephalitis virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 11021 / Sci species name: Eastern equine encephalitis virus / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
| Virus shell | Shell ID: 1 / Diameter: 660.0 Å |
-Macromolecule #1: Spike glycoprotein E1
| Macromolecule | Name: Spike glycoprotein E1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Eastern equine encephalitis virus (strain Florida 91-469) Eastern equine encephalitis virus (strain Florida 91-469)Strain: Florida 91-469 |
| Molecular weight | Theoretical: 47.938141 KDa |
| Recombinant expression | Organism:  Cricetinae gen. sp. (mammal) Cricetinae gen. sp. (mammal) |
| Sequence | String: YEHTAVMPNK VGIPYKALVE RPGYAPVHLQ IQLVNTRIIP STNLEYITCK YKTKVPSPVV KCCGATQCTS KPHPDYQCQV FTGVYPFMW GGAYCFCDTE NTQMSEAYVE RSEECSIDHA KAYKVHTGTV QAMVNITYGS VSWRSADVYV NGETPAKIGD A KLIIGPLS ...String: YEHTAVMPNK VGIPYKALVE RPGYAPVHLQ IQLVNTRIIP STNLEYITCK YKTKVPSPVV KCCGATQCTS KPHPDYQCQV FTGVYPFMW GGAYCFCDTE NTQMSEAYVE RSEECSIDHA KAYKVHTGTV QAMVNITYGS VSWRSADVYV NGETPAKIGD A KLIIGPLS SAWSPFDNKV VVYGHEVYNY DFPEYGTGKA GSFGDLQSRT STSNDLYANT NLKLQRPQAG IVHTPFTQAP SG FERWKRD KGAPLNDVAP FGCSIALEPL RAENCAVGSI PISIDIPDAA FTRISETPTV SDLECKITEC TYASDFGGIA TVA YKSSKA GNCPIHSPSG VAVIKENDVT LAESGSFTFH FSTANIHPAF KLQVCTSAVT CKGDCKPPKD HIVDYPAQHT ESFT SAISA TAWSWLKVLV GGTSAFIVLG LIATAVVALV LFFHRH UniProtKB: Structural polyprotein |
-Macromolecule #2: Spike glycoprotein E2
| Macromolecule | Name: Spike glycoprotein E2 / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Eastern equine encephalitis virus (strain Florida 91-469) Eastern equine encephalitis virus (strain Florida 91-469)Strain: Florida 91-469 |
| Molecular weight | Theoretical: 47.046953 KDa |
| Recombinant expression | Organism:  Cricetinae gen. sp. (mammal) Cricetinae gen. sp. (mammal) |
| Sequence | String: DLDTHFTQYK LARPYIADCP NCGHSRCDSP IAIEEVRGDA HAGVIRIQTS AMFGLKTDGV DLAYMSFMNG KTQKSIKIDN LHVRTSAPC SLVSHHGYYI LAQCPPGDTV TVGFHDGPNR HTCTVAHKVE FRPVGREKYR HPPEHGVELP CNRYTHKRAD Q GHYVEMHQ ...String: DLDTHFTQYK LARPYIADCP NCGHSRCDSP IAIEEVRGDA HAGVIRIQTS AMFGLKTDGV DLAYMSFMNG KTQKSIKIDN LHVRTSAPC SLVSHHGYYI LAQCPPGDTV TVGFHDGPNR HTCTVAHKVE FRPVGREKYR HPPEHGVELP CNRYTHKRAD Q GHYVEMHQ PGLVADHSLL SIHSAKVKIT VPSGAQVKYY CKCPDVREGI TSSDHTTTCT DVKQCRAYLI DNKKWVYNSG RL PRGEGDT FKGKLHVPFV PVKAKCIATL APEPLVEHKH RTLILHLHPD HPTLLTTRSL GSDANPTRQW IERPTTVNFT VTG EGLEYT WGNHPPKRVW AQESGEGNPH GWPHEVVVYY YNRYPLTTII GLCTCVAIIM VSCVTSVWLL CRTRNLCITP YKLA PNAQV PILLALLCCI KPTRA UniProtKB: Structural polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 5.5 Component:
Details: pH 5.5 | ||||||||||||||||||
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Details: 25 mA | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 298 K / Instrument: GATAN CRYOPLUNGE 3 / Details: blot for 3.3 seconds before plunging. | ||||||||||||||||||
| Details | Estimate E2 protein concentration by SDS-PAGE with BSA ranging from 0.1 to 1 microgram. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 100.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)