+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23739 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of MLL1-NCP (H3K4M) complex, mode02 | |||||||||
 Map data Map data | Mode02 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | MLL1-NCP / H3K4 methylation / TRANSFERASE / TRANSFERASE-DNA BINDING PROTEIN-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of DNA methylation-dependent heterochromatin formation / protein-cysteine methyltransferase activity / [histone H3]-lysine4 N-methyltransferase / histone H3K4 monomethyltransferase activity / response to potassium ion / unmethylated CpG binding / histone H3K4 trimethyltransferase activity / histone H3Q5ser reader activity / histone H3K4me1 reader activity / Epigenetic regulation of gene expression by MLL3 and MLL4 complexes ...negative regulation of DNA methylation-dependent heterochromatin formation / protein-cysteine methyltransferase activity / [histone H3]-lysine4 N-methyltransferase / histone H3K4 monomethyltransferase activity / response to potassium ion / unmethylated CpG binding / histone H3K4 trimethyltransferase activity / histone H3Q5ser reader activity / histone H3K4me1 reader activity / Epigenetic regulation of gene expression by MLL3 and MLL4 complexes / MLL3/4 complex / Set1C/COMPASS complex / MLL1/2 complex / definitive hemopoiesis / ATAC complex / regulation of short-term neuronal synaptic plasticity / NSL complex / histone H3K4 methyltransferase activity / Cardiogenesis / anterior/posterior pattern specification / T-helper 2 cell differentiation / embryonic hemopoiesis / Formation of WDR5-containing histone-modifying complexes / exploration behavior / minor groove of adenine-thymine-rich DNA binding / histone methyltransferase complex / hemopoiesis / MLL1 complex / regulation of embryonic development / regulation of cell division / histone acetyltransferase complex / membrane depolarization / cellular response to transforming growth factor beta stimulus / negative regulation of fibroblast proliferation / spleen development / transcription initiation-coupled chromatin remodeling / positive regulation of gluconeogenesis / homeostasis of number of cells within a tissue / Transferases; Transferring one-carbon groups; Methyltransferases / gluconeogenesis / post-embryonic development / skeletal system development / circadian regulation of gene expression / Deactivation of the beta-catenin transactivating complex / Formation of the beta-catenin:TCF transactivating complex / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / beta-catenin binding / visual learning / protein modification process / PKMTs methylate histone lysines / response to estrogen / Activation of anterior HOX genes in hindbrain development during early embryogenesis / RMTs methylate histone arginines / Transcriptional regulation of granulopoiesis / mitotic spindle / structural constituent of chromatin / heterochromatin formation / nucleosome / nucleosome assembly / HATs acetylate histones / RUNX1 regulates transcription of genes involved in differentiation of HSCs / Neddylation / MLL4 and MLL3 complexes regulate expression of PPARG target genes in adipogenesis and hepatic steatosis / protein-containing complex assembly / fibroblast proliferation / histone binding / methylation / transcription cis-regulatory region binding / regulation of cell cycle / protein heterodimerization activity / positive regulation of cell population proliferation / apoptotic process / DNA damage response / chromatin binding / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / positive regulation of DNA-templated transcription / nucleolus / negative regulation of transcription by RNA polymerase II / protein homodimerization activity / positive regulation of transcription by RNA polymerase II / DNA binding / zinc ion binding / nucleoplasm / identical protein binding / nucleus / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) / | |||||||||
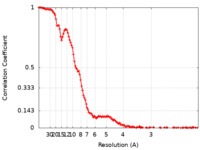
| Method | single particle reconstruction / cryo EM / Resolution: 4.02 Å | |||||||||
 Authors Authors | Park SH / Ayoub A | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Biochemistry / Year: 2022 Journal: Biochemistry / Year: 2022Title: Regulation of MLL1 Methyltransferase Activity in Two Distinct Nucleosome Binding Modes. Authors: Alex Ayoub / Sang Ho Park / Young-Tae Lee / Uhn-Soo Cho / Yali Dou /  Abstract: Cryo-EM structures of the KMT2A/MLL1 core complex bound on nucleosome core particles (NCPs) suggest unusual rotational dynamics of the MLL1 complex approaching its physiological substrate. However, ...Cryo-EM structures of the KMT2A/MLL1 core complex bound on nucleosome core particles (NCPs) suggest unusual rotational dynamics of the MLL1 complex approaching its physiological substrate. However, the functional implication of such dynamics remains unclear. Here, we show that the MLL1 core complex also shows high rotational dynamics bound on the NCP carrying the catalytically inert histone H3 lysine 4 to methionine (K4M) mutation. There are two major binding modes of the MLL1 complex on the NCP. Importantly, disruption of only one of the binding modes compromised the overall MLL1 activity in an NCP-specific manner. We propose that the MLL1 core complex probably exists in an equilibrium of poised and active binding modes. The high rotational dynamics of the MLL1 complex on the NCP is a feature that can be exploited for loci-specific regulation of H3K4 methylation in higher eukaryotes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23739.map.gz emd_23739.map.gz | 14.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23739-v30.xml emd-23739-v30.xml emd-23739.xml emd-23739.xml | 26.8 KB 26.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23739_fsc.xml emd_23739_fsc.xml | 16.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_23739.png emd_23739.png | 64.1 KB | ||
| Filedesc metadata |  emd-23739.cif.gz emd-23739.cif.gz | 8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23739 http://ftp.pdbj.org/pub/emdb/structures/EMD-23739 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23739 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23739 | HTTPS FTP |
-Related structure data
| Related structure data |  7mbnMC  7mbmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23739.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23739.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mode02 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
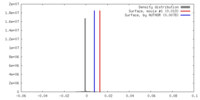
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nu...
+Supramolecule #1: MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nu...
+Supramolecule #2: RbBP5, WDR5, SET1, and ASH2L
+Supramolecule #3: nucleosome
+Macromolecule #1: Retinoblastoma-binding protein 5
+Macromolecule #2: WD repeat-containing protein 5
+Macromolecule #3: Histone-lysine N-methyltransferase 2A
+Macromolecule #4: Set1/Ash2 histone methyltransferase complex subunit ASH2
+Macromolecule #5: Histone H3
+Macromolecule #6: Histone H4
+Macromolecule #7: Histone H2A
+Macromolecule #8: Histone H2B 1.1
+Macromolecule #9: DNA (146-MER)
+Macromolecule #10: DNA (146-MER)
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 53.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7mbn: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)