[English] 日本語
 Yorodumi
Yorodumi- EMDB-23274: PF 06882961 bound to the glucagon-like peptide-1 receptor (GLP-1R... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23274 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | PF 06882961 bound to the glucagon-like peptide-1 receptor (GLP-1R):Gs complex | ||||||||||||||||||
 Map data Map data | main map, consensus refinement, post-processed | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | GPCR / small molecule agonist / MEMBRANE PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationglucagon-like peptide 1 receptor activity / glucagon receptor activity / positive regulation of blood pressure / hormone secretion / post-translational protein targeting to membrane, translocation / response to psychosocial stress / regulation of heart contraction / peptide hormone binding / PKA activation in glucagon signalling / developmental growth ...glucagon-like peptide 1 receptor activity / glucagon receptor activity / positive regulation of blood pressure / hormone secretion / post-translational protein targeting to membrane, translocation / response to psychosocial stress / regulation of heart contraction / peptide hormone binding / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / D1 dopamine receptor binding / intracellular transport / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity / Hedgehog 'off' state / negative regulation of blood pressure / adenylate cyclase-activating adrenergic receptor signaling pathway / regulation of insulin secretion / cellular response to glucagon stimulus / adenylate cyclase activator activity / trans-Golgi network membrane / negative regulation of inflammatory response to antigenic stimulus / bone development / platelet aggregation / cognition / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / Adrenaline,noradrenaline inhibits insulin secretion / sensory perception of smell / Vasopressin regulates renal water homeostasis via Aquaporins / transmembrane signaling receptor activity / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / positive regulation of cold-induced thermogenesis / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / positive regulation of cytosolic calcium ion concentration / G protein activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / learning or memory / cell surface receptor signaling pathway / Extra-nuclear estrogen signaling / cell population proliferation / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / synapse / GTP binding / protein-containing complex binding / signal transduction / extracellular exosome / metal ion binding / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
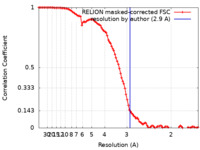
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | ||||||||||||||||||
 Authors Authors | Belousoff MJ / Johnson RM | ||||||||||||||||||
| Funding support |  Australia, Australia,  Japan, 5 items Japan, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2021 Journal: Structure / Year: 2021Title: Evolving cryo-EM structural approaches for GPCR drug discovery. Authors: Xin Zhang / Rachel M Johnson / Ieva Drulyte / Lingbo Yu / Abhay Kotecha / Radostin Danev / Denise Wootten / Patrick M Sexton / Matthew J Belousoff /    Abstract: G protein-coupled receptors (GPCRs) are the largest class of cell surface drug targets. Advances in stabilization of GPCR:transducer complexes, together with improvements in cryoelectron microscopy ...G protein-coupled receptors (GPCRs) are the largest class of cell surface drug targets. Advances in stabilization of GPCR:transducer complexes, together with improvements in cryoelectron microscopy (cryo-EM) have recently been applied to structure-assisted drug design for GPCR agonists. Nonetheless, limitations in the commercial application of these approaches, including the use of nanobody 35 (Nb35) to aid complex stabilization and the high cost of 300 kV imaging, have restricted broad application of cryo-EM in drug discovery. Here, using the PF 06882961-bound GLP-1R as exemplar, we validated the formation of stable complexes with a modified Gs protein in the absence of Nb35. In parallel, we compare 200 versus 300 kV image acquisition using a Falcon 4 or K3 direct electron detector. Moreover, the 200 kV Glacios-Falcon 4 yielded a 3.2 Å map with clear density for bound drug and multiple structurally ordered waters. Our work paves the way for broader commercial application of cryo-EM for GPCR drug discovery. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23274.map.gz emd_23274.map.gz | 85.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23274-v30.xml emd-23274-v30.xml emd-23274.xml emd-23274.xml | 30.4 KB 30.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23274_fsc.xml emd_23274_fsc.xml | 10.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_23274.png emd_23274.png | 38.6 KB | ||
| Filedesc metadata |  emd-23274.cif.gz emd-23274.cif.gz | 7.5 KB | ||
| Others |  emd_23274_additional_1.map.gz emd_23274_additional_1.map.gz emd_23274_additional_2.map.gz emd_23274_additional_2.map.gz emd_23274_additional_3.map.gz emd_23274_additional_3.map.gz emd_23274_half_map_1.map.gz emd_23274_half_map_1.map.gz emd_23274_half_map_2.map.gz emd_23274_half_map_2.map.gz | 84.6 MB 59.5 MB 85.3 MB 71.7 MB 71.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23274 http://ftp.pdbj.org/pub/emdb/structures/EMD-23274 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23274 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23274 | HTTPS FTP |
-Related structure data
| Related structure data |  7lciMC  7lcjC  7lckC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23274.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23274.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map, consensus refinement, post-processed | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
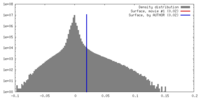
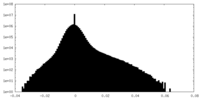
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: receptor focussed refinement, post-processed, resampled on consensus map...
| File | emd_23274_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | receptor focussed refinement, post-processed, resampled on consensus map | ||||||||||||
| Projections & Slices |
| ||||||||||||
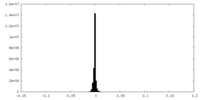
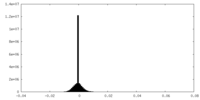
| Density Histograms |
-Additional map: local resolution filtered consensus map
| File | emd_23274_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local resolution filtered consensus map | ||||||||||||
| Projections & Slices |
| ||||||||||||
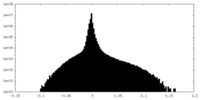
| Density Histograms |
-Additional map: g-protein focussed refinment
| File | emd_23274_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | g-protein focussed refinment | ||||||||||||
| Projections & Slices |
| ||||||||||||
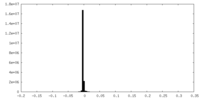
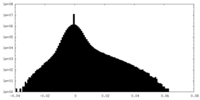
| Density Histograms |
-Half map: half map1: consensus
| File | emd_23274_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map1: consensus | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map2: consensus
| File | emd_23274_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map2: consensus | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : GLP-1:Gs complex with small molecule agonist bound (PF-06882961)
| Entire | Name: GLP-1:Gs complex with small molecule agonist bound (PF-06882961) |
|---|---|
| Components |
|
-Supramolecule #1: GLP-1:Gs complex with small molecule agonist bound (PF-06882961)
| Supramolecule | Name: GLP-1:Gs complex with small molecule agonist bound (PF-06882961) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Glucagon-like peptide 1 receptor
| Macromolecule | Name: Glucagon-like peptide 1 receptor / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 56.748418 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDLEVLFQG PARPQGATVS LWETVQKWRE YRRQCQRSLT EDPPPATDLF CNRTFDEYAC WPDGEPGSF VNVSCPWYLP WASSVPQGHV YRFCTAEGLW LQKDNSSLPW RDLSECEESK RGERSSPEEQ LLFLYIIYTV G YALSFSAL ...String: MKTIIALSYI FCLVFADYKD DDDLEVLFQG PARPQGATVS LWETVQKWRE YRRQCQRSLT EDPPPATDLF CNRTFDEYAC WPDGEPGSF VNVSCPWYLP WASSVPQGHV YRFCTAEGLW LQKDNSSLPW RDLSECEESK RGERSSPEEQ LLFLYIIYTV G YALSFSAL VIASAILLGF RHLHCTRNYI HLNLFASFIL RALSVFIKDA ALKWMYSTAA QQHQWDGLLS YQDSLSCRLV FL LMQYCVA ANYYWLLVEG VYLYTLLAFS VFSEQWIFRL YVSIGWGVPL LFVVPWGIVK YLYEDEGCWT RNSNMNYWLI IRL PILFAI GVNFLIFVRV ICIVVSKLKA NLMCKTDIKC RLAKSTLTLI PLLGTHEVIF AFVMDEHARG TLRFIKLFTE LSFT SFQGL MVAILYCFVN NEVQLEFRKS WERWRLEHLH IQRDSSMKPL KCPTSSLSSG ATAGSSMYTA TCQASCSPAG LEVLF QGPH HHHHHHH UniProtKB: Glucagon-like peptide 1 receptor |
-Macromolecule #2: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.699434 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKNTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE ...String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKNTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE YQLIDCAQYF LDKIDVIKQA DYVPSDQDLL RCRVLTSGIF ETKFQVDKVN FHMFDVGAQR DERRKWIQCF ND VTAIIFV VASSSYNMVI REDNQTNRLQ AALKLFDSIW NNKWLRDTSV ILFLNKQDLL AEKVLAGKSK IEDYFPEFAR YTT PEDATP EPGEDPRVTR AKYFIRDEFL RISTASGDGR HYCYPHFTCS VDTENIRRVF NDCRDIIQRM HLRQYELL UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.413863 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: QSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL ...String: QSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD INAICFFPNG NA FATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAGHDNRVSC LGV TDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 6.375332 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: NTASIAQARK LVEQLKMEAN IDRIKVSKAA ADLMAYCEAH AKEDPLLTPV PASENPFR UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: 2-[(4-{6-[(4-cyano-2-fluorophenyl)methoxy]pyridin-2-yl}piperidin-...
| Macromolecule | Name: 2-[(4-{6-[(4-cyano-2-fluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-{[(2S)-oxetan-2-yl]methyl}-1H-benzimidazole-6-carboxylic acid type: ligand / ID: 5 / Number of copies: 1 / Formula: UK4 |
|---|---|
| Molecular weight | Theoretical: 555.599 Da |
| Chemical component information |  ChemComp-UK4: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 20 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 61.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)