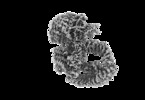
登録情報 データベース : EMDB / ID : EMD-23027タイトル Cryo-EM structure of the human SAGA coactivator complex (TRRAP, core) LocSpiral filtered map 複合体 : human SAGA複合体 : human SAGA複合体 : STAGA complex 65 subunit gammaリガンド : x 1種 / / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Homo sapiens (ヒト) / unclassified Rhodococcus (バクテリア)手法 / / 解像度 : 2.93 Å Herbst DA / Esbin MN 資金援助 Organization Grant number 国 Howard Hughes Medical Institute (HHMI) CC30250 National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) R35GM127018 European Molecular Biology Organization (EMBO) ALTF 1002-2018 European Union Swiss National Science Foundation P2BSP3_181878 Howard Hughes Medical Institute (HHMI) CC34430 National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) T32GM098218
ジャーナル : Nat Struct Mol Biol / 年 : 2021タイトル : Structure of the human SAGA coactivator complex.著者 : Dominik A Herbst / Meagan N Esbin / Robert K Louder / Claire Dugast-Darzacq / Gina M Dailey / Qianglin Fang / Xavier Darzacq / Robert Tjian / Eva Nogales / 要旨 : The SAGA complex is a regulatory hub involved in gene regulation, chromatin modification, DNA damage repair and signaling. While structures of yeast SAGA (ySAGA) have been reported, there are ... The SAGA complex is a regulatory hub involved in gene regulation, chromatin modification, DNA damage repair and signaling. While structures of yeast SAGA (ySAGA) have been reported, there are noteworthy functional and compositional differences for this complex in metazoans. Here we present the cryogenic-electron microscopy (cryo-EM) structure of human SAGA (hSAGA) and show how the arrangement of distinct structural elements results in a globally divergent organization from that of yeast, with a different interface tethering the core module to the TRRAP subunit, resulting in a dramatically altered geometry of functional elements and with the integration of a metazoan-specific splicing module. Our hSAGA structure reveals the presence of an inositol hexakisphosphate (InsP) binding site in TRRAP and an unusual property of its pseudo-(Ψ)PIKK. Finally, we map human disease mutations, thus providing the needed framework for structure-guided drug design of this important therapeutic target for human developmental diseases and cancer. 履歴 登録 2020年11月24日 - ヘッダ(付随情報) 公開 2021年11月10日 - マップ公開 2021年11月10日 - 更新 2024年5月29日 - 現状 2024年5月29日 処理サイト : RCSB / 状態 : 公開
すべて表示 表示を減らす
 万見
万見 データを開く
データを開く 基本情報
基本情報 マップデータ
マップデータ 試料
試料 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト) /
Homo sapiens (ヒト) /  unclassified Rhodococcus (バクテリア)
unclassified Rhodococcus (バクテリア) データ登録者
データ登録者 米国, European Union,
米国, European Union,  スイス, 6件
スイス, 6件  引用
引用 ジャーナル: Nat Struct Mol Biol / 年: 2021
ジャーナル: Nat Struct Mol Biol / 年: 2021

 構造の表示
構造の表示 ムービービューア
ムービービューア SurfView
SurfView Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク emd_23027.map.gz
emd_23027.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-23027-v30.xml
emd-23027-v30.xml emd-23027.xml
emd-23027.xml EMDBヘッダ
EMDBヘッダ emd_23027_fsc.xml
emd_23027_fsc.xml FSCデータファイル
FSCデータファイル emd_23027.png
emd_23027.png emd_23027_msk_1.map
emd_23027_msk_1.map マスクマップ
マスクマップ emd-23027.cif.gz
emd-23027.cif.gz emd_23027_additional_1.map.gz
emd_23027_additional_1.map.gz emd_23027_additional_2.map.gz
emd_23027_additional_2.map.gz emd_23027_additional_3.map.gz
emd_23027_additional_3.map.gz emd_23027_half_map_1.map.gz
emd_23027_half_map_1.map.gz emd_23027_half_map_2.map.gz
emd_23027_half_map_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-23027
http://ftp.pdbj.org/pub/emdb/structures/EMD-23027 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23027
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23027 emd_23027_validation.pdf.gz
emd_23027_validation.pdf.gz EMDB検証レポート
EMDB検証レポート emd_23027_full_validation.pdf.gz
emd_23027_full_validation.pdf.gz emd_23027_validation.xml.gz
emd_23027_validation.xml.gz emd_23027_validation.cif.gz
emd_23027_validation.cif.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23027
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23027 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23027
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23027 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_23027.map.gz / 形式: CCP4 / 大きさ: 178 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_23027.map.gz / 形式: CCP4 / 大きさ: 178 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) emd_23027_msk_1.map
emd_23027_msk_1.map 試料の構成要素
試料の構成要素 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN
 画像解析
画像解析
 ムービー
ムービー コントローラー
コントローラー



















 Z
Z Y
Y X
X