+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22166 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Assembly intermediates of orthoreovirus captured in the cell | ||||||||||||
 Map data Map data | DLP-pen-5 fold | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | orthoreovirus / VIRUS LIKE PARTICLE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationicosahedral viral capsid / host cell surface binding / viral inner capsid / symbiont-mediated suppression of host PKR/eIFalpha signaling / viral outer capsid / symbiont entry into host cell via permeabilization of host membrane / host cell endoplasmic reticulum / host cell mitochondrion / 7-methylguanosine mRNA capping / protein serine/threonine kinase inhibitor activity ...icosahedral viral capsid / host cell surface binding / viral inner capsid / symbiont-mediated suppression of host PKR/eIFalpha signaling / viral outer capsid / symbiont entry into host cell via permeabilization of host membrane / host cell endoplasmic reticulum / host cell mitochondrion / 7-methylguanosine mRNA capping / protein serine/threonine kinase inhibitor activity / viral life cycle / viral capsid / regulation of translation / mRNA guanylyltransferase activity / mRNA guanylyltransferase / mRNA (guanine-N7)-methyltransferase / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / RNA helicase activity / symbiont-mediated suppression of host innate immune response / RNA helicase / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / GTP binding / host cell plasma membrane / structural molecule activity / ATP hydrolysis activity / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | ||||||||||||
| Biological species |  Mammalian orthoreovirus 3 Dearing / Mammalian orthoreovirus 3 Dearing /  Reovirus type 1 (strain Lang) Reovirus type 1 (strain Lang) | ||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 6.5 Å | ||||||||||||
 Authors Authors | Sutton G / Sun DP | ||||||||||||
| Funding support |  United Kingdom, United Kingdom,  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Assembly intermediates of orthoreovirus captured in the cell. Authors: Geoff Sutton / Dapeng Sun / Xiaofeng Fu / Abhay Kotecha / Corey W Hecksel / Daniel K Clare / Peijun Zhang / David I Stuart / Mark Boyce /    Abstract: Traditionally, molecular assembly pathways for viruses are inferred from high resolution structures of purified stable intermediates, low resolution images of cell sections and genetic approaches. ...Traditionally, molecular assembly pathways for viruses are inferred from high resolution structures of purified stable intermediates, low resolution images of cell sections and genetic approaches. Here, we directly visualise an unsuspected 'single shelled' intermediate for a mammalian orthoreovirus in cryo-preserved infected cells, by cryo-electron tomography of cellular lamellae. Particle classification and averaging yields structures to 5.6 Å resolution, sufficient to identify secondary structural elements and produce an atomic model of the intermediate, comprising 120 copies each of protein λ1 and σ2. This λ1 shell is 'collapsed' compared to the mature virions, with molecules pushed inwards at the icosahedral fivefolds by ~100 Å, reminiscent of the first assembly intermediate of certain prokaryotic dsRNA viruses. This supports the supposition that these viruses share a common ancestor, and suggests mechanisms for the assembly of viruses of the Reoviridae. Such methodology holds promise for dissecting the replication cycle of many viruses. #1:  Journal: To Be Published Journal: To Be PublishedTitle: Assembly intermediates of orthoreovirus captured in the cell Authors: Sutton G / Sun DP / Fu XF / Kotecha A / Hecksel GW / Clare DK / Zhang P / Stuart D / Boyce M | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22166.map.gz emd_22166.map.gz | 19.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22166-v30.xml emd-22166-v30.xml emd-22166.xml emd-22166.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22166.png emd_22166.png | 94.2 KB | ||
| Filedesc metadata |  emd-22166.cif.gz emd-22166.cif.gz | 7.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22166 http://ftp.pdbj.org/pub/emdb/structures/EMD-22166 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22166 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22166 | HTTPS FTP |
-Related structure data
| Related structure data |  6xf8MC  6ztzMC  6xf7C  6ztsC  6ztyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22166.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22166.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DLP-pen-5 fold | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : reovirus SLP
| Entire | Name: reovirus SLP |
|---|---|
| Components |
|
-Supramolecule #1: reovirus SLP
| Supramolecule | Name: reovirus SLP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Mammalian orthoreovirus 3 Dearing Mammalian orthoreovirus 3 Dearing |
-Macromolecule #1: Outer capsid protein mu-1
| Macromolecule | Name: Outer capsid protein mu-1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Reovirus type 1 (strain Lang) / Strain: Lang Reovirus type 1 (strain Lang) / Strain: Lang |
| Molecular weight | Theoretical: 68.568648 KDa |
| Sequence | String: PGGVPWIAIG DETSVTSPGA LRRMTSKDIP ETAIINTDNS SGAVPSESAL VPYNDEPLVV VTEHAIANFT KAEMALEFNR EFLDKLRVL SVSPKYSDLL TYVDCYVGVS ARQALNNFQK QVPVITPTRQ TMYVDSIQAA LKALEKWEID LRVAQTLLPT N VPIGEVSC ...String: PGGVPWIAIG DETSVTSPGA LRRMTSKDIP ETAIINTDNS SGAVPSESAL VPYNDEPLVV VTEHAIANFT KAEMALEFNR EFLDKLRVL SVSPKYSDLL TYVDCYVGVS ARQALNNFQK QVPVITPTRQ TMYVDSIQAA LKALEKWEID LRVAQTLLPT N VPIGEVSC PMQSVVKLLD DQLPDDSLIR RYPKEAAVAL AKRNGGIQWM DVSEGTVMNE AVNAVAASAL APSASAPPLE EK SKLTEQA MDLVTAAEPE IIASLVPVPA PVFAIPPKPA DYNVRTLKID EATWLRMIPK TMGTLFQIQV TDNTGTNWHF NLR GGTRVV NLDQIAPMRF VLDLGGKSYK ETSWDPNGKK VGFIVFQSKI PFELWTAASQ IGQATVVNYV QLYAEDSSFT AQSI IATTS LAYNYEPEQL NKTDPEMNYY LLATFIDSAA ITPTNMTQPD VWDALLTMSP LSAGEVTVKG AVVSEVVPAE LIGSY TPES LNASLPNDAA RCMIDRASKI AEAIKIDDDA GPDEYSPNSV PIQGQLAISQ LETGYGVRIF NPKGILSKIA SRAMQA FIG DPSTIITQAA PVLSDKNNWI ALAQGVKTSL RTKSLSAGVK TAVSKLSSSE SIQNWTQGFL DKVSTHFPAP UniProtKB: Outer capsid protein mu-1 |
-Macromolecule #2: Outer capsid protein sigma-3
| Macromolecule | Name: Outer capsid protein sigma-3 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Reovirus type 1 (strain Lang) / Strain: Lang Reovirus type 1 (strain Lang) / Strain: Lang |
| Molecular weight | Theoretical: 41.237117 KDa |
| Sequence | String: MEVCLPNGHQ IVDLINNAFE GRVSIYSAQE GWDKTISAQP DMMVCGGAVV CMHCLGVVGS LQRKLKHLPH HRCNQQIRHQ DYVDVQFAD RVTAHWKRGM LSFVCQMHAM MNDVSPEDLD RVRTEGGSLV ELNWLQVDPN SMFRSIHSSW TDPLQVVDDL D TKLDQYWT ...String: MEVCLPNGHQ IVDLINNAFE GRVSIYSAQE GWDKTISAQP DMMVCGGAVV CMHCLGVVGS LQRKLKHLPH HRCNQQIRHQ DYVDVQFAD RVTAHWKRGM LSFVCQMHAM MNDVSPEDLD RVRTEGGSLV ELNWLQVDPN SMFRSIHSSW TDPLQVVDDL D TKLDQYWT ALNLMIDSSD LVPNFMMRDP SHAFNGVRLE GDARQTQFSR TFDSRSSLEW GVMVYDYSEL EHDPSKGRAY RK ELVTPAR DFGHFGLSHY SRATTPILGK MPAVFSGMLT GNCKMYPFIK GTAKLKTVRK LVDSVNHAWG VEKIRYALGP GGM TGWYNR TMQQAPIVLT PAALTMFSDT TKFGDLDYPV MIGDPMILG UniProtKB: Outer capsid protein sigma-3 |
-Macromolecule #3: Inner capsid protein sigma-2
| Macromolecule | Name: Inner capsid protein sigma-2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Reovirus type 1 (strain Lang) / Strain: Lang Reovirus type 1 (strain Lang) / Strain: Lang |
| Molecular weight | Theoretical: 47.024016 KDa |
| Sequence | String: ARAAFLFKTV GFGGLQNVPI NDELSSHLLR AGNSPWQLTQ FLDWISLGRG LATSALVPTA GSRYYQMSCL LSGTLQIPFR PNHRWGDIR FLRLVWSAPT LDGLVVAPPQ VLAQPALQAQ ADRVYDCDDY PFLARDPRFK HRVYQQLSAV TLLNLTGFGP I SYVRVDED ...String: ARAAFLFKTV GFGGLQNVPI NDELSSHLLR AGNSPWQLTQ FLDWISLGRG LATSALVPTA GSRYYQMSCL LSGTLQIPFR PNHRWGDIR FLRLVWSAPT LDGLVVAPPQ VLAQPALQAQ ADRVYDCDDY PFLARDPRFK HRVYQQLSAV TLLNLTGFGP I SYVRVDED MWSGDVNQLL MNYFGHTFAE IAYTLCQASA NRPWEHDGTY ARMTQIILSL FWLSYVGVIH QQNTYRTFYF QC NRRGDAA EVWILSCSLN HSAQIRPGNR SLFVMPTSPD WNMDVNLILS STLTGCLCSG SQLPLIDNNS VPAVSRNIHG WTG RAGNQL HGFQVRRMVT EFCDRLRRDG VMTQAQQNQI EALADQTQQF KRDKLEAWAR EDDQYNQANP NSTMFRTKPF TNAQ WGRGN TGATSAAIAA LI UniProtKB: Inner capsid protein sigma-2 |
-Macromolecule #4: Inner capsid protein lambda-1
| Macromolecule | Name: Inner capsid protein lambda-1 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO / EC number: RNA helicase |
|---|---|
| Source (natural) | Organism:  Reovirus type 1 (strain Lang) / Strain: Lang Reovirus type 1 (strain Lang) / Strain: Lang |
| Molecular weight | Theoretical: 119.020562 KDa |
| Sequence | String: QRHITEFISS WQNHPIVQVS ADVENKKTAQ LLHADTPRLV TWDAGLCTSF KIVPIVPAQV PQDVLAYTFF TSSYAIQSPF PEAAVSRIV VHTRWASNVD FDRDSSVIMA PPTENNIHLF KQLLNTETLS VRGANPLMFR ANVLHMLLEF VLDNLYLNRH T GFSQDHTP ...String: QRHITEFISS WQNHPIVQVS ADVENKKTAQ LLHADTPRLV TWDAGLCTSF KIVPIVPAQV PQDVLAYTFF TSSYAIQSPF PEAAVSRIV VHTRWASNVD FDRDSSVIMA PPTENNIHLF KQLLNTETLS VRGANPLMFR ANVLHMLLEF VLDNLYLNRH T GFSQDHTP FTEGANLRSL PGPDAEKWYS IMYPTRMGTP NVSKICNFVA SCVRNRVGRF DRAQMMNGAM SEWVDVFETS DA LTVSIRG RWMARLARMN INPTEIEWAL TECAQGYVTV TSPYAPSVNR LMPYRISNAE RQISQIIRIM NIGNNATVIQ PVL QDISVL LQRISPLQID PTIISNTMST VSESTTQTLS PASSILGKLR PSNSDFSSFR VALAGWLYNG VVTTVIDDSS YPKD GGSVT SLENLWDFFI LALALPLTTD PCAPVKAFMT LANMMVGFET IPMDNQIYTQ SRRASAFSTP HTWPRCFMNI QLISP IDAP ILRQWAEIIH RYWPNPSQIR YGAPNVFGSA NLFTPPEVLL LPIDHQPANV TTPTLDFTNE LTNWRARVCE LMKNLV DNQ RYQPGWTQSL VSSMRGTLDK LKLIKSMTPM YLQQLAPVEL AVIAPMLPFP PFQVPYVRLD RDRVPTMVGV TRQSRDT IT QPALSLSTTN TTVGVPLALD ARAITVALLS GKYPPDLVTN VWYADAIYPM YADTEVFSNL QRDMITCEAV QTLVTLVA Q ISETQYPVDR YLDWIPSLRA SAATAATFAE WVNTSMKTAF DLSDMLLEPL LSGDPRMTQL AIQYQQYNGR TFNVIPEMP GSVIADCVQL TAEVFNHEYN LFGIARGDII IGRVQSTHLW SPLAPPPDLV FDRDTPGVHI FGRDCRISFG MNGAAPMIRD ETGMMVPFE GNWIFPLALW QMNTRYFNQQ FDAWIKTGEL RIRIEMGAYP YMLHYYDPRQ YANAWNLTSA WLEEITPTSI P SVPFMVPI SSDHDISSAP AVQYIISTEY NDRSLFCTNS SSPQTIAGPD KHIPVERYNI LTNPDAPPTQ IQLPEVVDLY NV VTRYAYE TPPITAVVMG VP UniProtKB: Inner capsid protein lambda-1 |
-Macromolecule #5: mRNA (guanine-N(7)-)-methyltransferase
| Macromolecule | Name: mRNA (guanine-N(7)-)-methyltransferase / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO / EC number: mRNA (guanine-N7)-methyltransferase |
|---|---|
| Source (natural) | Organism:  Reovirus type 1 (strain Lang) / Strain: Lang Reovirus type 1 (strain Lang) / Strain: Lang |
| Molecular weight | Theoretical: 143.967562 KDa |
| Sequence | String: ANVWGVRLAD SLSSPTIETR TRQYTLHDLC SDLDANPGRE PWKPLRNQRT NNIVAVQLFR PLQGLVLDTQ LYGFPGAFDD WERFMREKL RVLKYEVLRI YPISNYSNEH VNVFVANALV GAFLSNQAFY DLLPLLIIND TMIGDLLGTG ASLSQFFQSH G DVLEVAAG ...String: ANVWGVRLAD SLSSPTIETR TRQYTLHDLC SDLDANPGRE PWKPLRNQRT NNIVAVQLFR PLQGLVLDTQ LYGFPGAFDD WERFMREKL RVLKYEVLRI YPISNYSNEH VNVFVANALV GAFLSNQAFY DLLPLLIIND TMIGDLLGTG ASLSQFFQSH G DVLEVAAG RKYLQMENYS NDDDDPPLFA KDLSDYAKAF YSDTYEVLDR FFWTHDSSAG VLVHYDKPTN GHHYLLGTLT QM VSAPPYI INATDAMLLE SCLEQFSANV RARPAQPVTR LDQCYHLRWG AQYVGEDSLT YRLGVLSLLA TNGYQLARPI PRQ LTNRWL SSFVSQIMSD GVNETPLWPQ ERYVQIAYDS PSVVDGATQY GYVRKNQLRL GMRISALQSL SDTPSPVQWL PQYT IDQAA MDEGDLMVSR LTQLPLRPDY GNIWVGDALS YYVDYNRSHR VVLSSELPQL PDTYFDGDEQ YGRSLFSLAR KIGDR SLVK DTAVLKHAYQ AIDPNTGKEY LRSRQSVAYF GASAGHSGAD QPLVIEPWIQ GKISGVPPPS SVRQFGYDVA RGAIVD LAR PFPSGDYQFV YSDVDQVVDG HDDLSISSGL VESLLSSCMH ATAPGGSFVV KINFPTRPVW HYIEQKILPN ITSYMLI KP FVTNNVELFF VAFGVHQHSS LTWTSGVYFF LVDHFYRYET LSTISRQLPS FGYVDDGSSV TGIETISIEN PGFSNMTQ A ARIGISGLCA NVGNARKSIA IYESHGARVL TITSRRSPAS ARRKSRLRYL PLIDPRSLEV QARTILPADP VLFENVSGA SPHVCLTMMY NFEVSSAVYD GDVVLDLGTG PEAKILELIP ATSPVTCVDI RPTAQPSGCW NVRTTFLELD YLSDGWITGV RGDIVTCML SLGAAAAGKS MTFDAAFQQL IKVLSKSTAN VVLVQVNCPT DVVRSIKGYL EIDSTNKRYR FPKFGRDEPY S DMDALEKI CRTAWPNCSI TWVPLSYDLR WTRLALLEST TLSSASIRIA ELMYKYMPIM RIDIHGLPME KRGNFIVGQN CS LVIPGFN AQDVFNCYFN SALAFSTEDV NAAMIPQVSA QFDATKGEWT LDMVFSDAGI YTMQALVGSN ANPVSLGSFV VDS PDVDIT DAWPAQLDFT IAGTDVDITV NPYYRLMTFV RIDGQWQIAN PDKFQFFSSA SGTLVMNVKL DIADKYLLYY IRDV QSRDV GFYIQHPLQL LNTITLPTNE DLFLSAPDMR EWAVKESGNT ICILNSQGFV LPQDWDVLTD TISWSPSIPT YIVPP GDYT LTPL UniProtKB: Outer capsid protein lambda-2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C5 (5 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 6.5 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: emClarity / Number subtomograms used: 625 |
|---|---|
| Extraction | Number tomograms: 4 / Number images used: 625 |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller






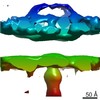

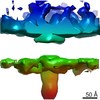

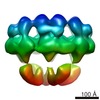







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















