+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20283 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Active State of Manduca sexta soluble Guanylate Cyclase | |||||||||
 Map data Map data | Map of the Active Ms sGC dimer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nitric oxide / cyclase / H-NOX / stimulator / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationguanylate cyclase complex, soluble / guanylate cyclase / guanylate cyclase activity / response to oxygen levels / : / heme binding / GTP binding Similarity search - Function | |||||||||
| Biological species |  Manduca sexta (tobacco hornworm) Manduca sexta (tobacco hornworm) | |||||||||
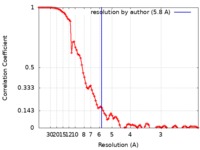
| Method | single particle reconstruction / cryo EM / Resolution: 5.8 Å | |||||||||
 Authors Authors | Yokom AL / Horst BG | |||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: Allosteric activation of the nitric oxide receptor soluble guanylate cyclase mapped by cryo-electron microscopy. Authors: Benjamin G Horst / Adam L Yokom / Daniel J Rosenberg / Kyle L Morris / Michal Hammel / James H Hurley / Michael A Marletta /  Abstract: Soluble guanylate cyclase (sGC) is the primary receptor for nitric oxide (NO) in mammalian nitric oxide signaling. We determined structures of full-length sGC in both inactive and active states ...Soluble guanylate cyclase (sGC) is the primary receptor for nitric oxide (NO) in mammalian nitric oxide signaling. We determined structures of full-length sGC in both inactive and active states using cryo-electron microscopy. NO and the sGC-specific stimulator YC-1 induce a 71° rotation of the heme-binding β H-NOX and PAS domains. Repositioning of the β H-NOX domain leads to a straightening of the coiled-coil domains, which, in turn, use the motion to move the catalytic domains into an active conformation. YC-1 binds directly between the β H-NOX domain and the two CC domains. The structural elongation of the particle observed in cryo-EM was corroborated in solution using small angle X-ray scattering (SAXS). These structures delineate the endpoints of the allosteric transition responsible for the major cyclic GMP-dependent physiological effects of NO. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20283.map.gz emd_20283.map.gz | 1.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20283-v30.xml emd-20283-v30.xml emd-20283.xml emd-20283.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20283_fsc.xml emd_20283_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_20283.png emd_20283.png | 36.3 KB | ||
| Filedesc metadata |  emd-20283.cif.gz emd-20283.cif.gz | 5.9 KB | ||
| Others |  emd_20283_half_map_1.map.gz emd_20283_half_map_1.map.gz emd_20283_half_map_2.map.gz emd_20283_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20283 http://ftp.pdbj.org/pub/emdb/structures/EMD-20283 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20283 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20283 | HTTPS FTP |
-Related structure data
| Related structure data |  6patMC  6pasC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20283.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20283.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of the Active Ms sGC dimer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.137 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half Map 2 of the Active Ms sGC
| File | emd_20283_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 of the Active Ms sGC | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 1 of the Active Ms sGC dimer
| File | emd_20283_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 of the Active Ms sGC dimer | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Active state of heterodimeric Ms sGC
| Entire | Name: Active state of heterodimeric Ms sGC |
|---|---|
| Components |
|
-Supramolecule #1: Active state of heterodimeric Ms sGC
| Supramolecule | Name: Active state of heterodimeric Ms sGC / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Manduca sexta (tobacco hornworm) Manduca sexta (tobacco hornworm) |
-Macromolecule #1: Soluble guanylyl cyclase alpha-1 subunit
| Macromolecule | Name: Soluble guanylyl cyclase alpha-1 subunit / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Manduca sexta (tobacco hornworm) Manduca sexta (tobacco hornworm) |
| Molecular weight | Theoretical: 78.598727 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 2 RZ-2014 (butterflies/moths) Spodoptera aff. frugiperda 2 RZ-2014 (butterflies/moths) |
| Sequence | String: MTCPFRRASS QHQFANGGSS APKKPEFRSR TSSVHLTGPE EEDGERNTLT LKHMSEALQL LTAPSNECLH AAVTSLTKNQ SDHYHKYNC LRRLPDDVKT CRNYAYLQEI YDAVRATDSV NTKDFMAKLG EYLILTAFSH NCRLERAFKC LGTNLTEFLT T LDSVHDVL ...String: MTCPFRRASS QHQFANGGSS APKKPEFRSR TSSVHLTGPE EEDGERNTLT LKHMSEALQL LTAPSNECLH AAVTSLTKNQ SDHYHKYNC LRRLPDDVKT CRNYAYLQEI YDAVRATDSV NTKDFMAKLG EYLILTAFSH NCRLERAFKC LGTNLTEFLT T LDSVHDVL HDQDTPLKDE TMEYEANFVC TTSQEGKIQL HLTTESEPVA YLLVGSLKAI AKRLYDTQTD IRLRSYTNDP RR FRYEINA VPLHQKSKED SCELVNEAAS VATSTKVTDL KIGVASFCKA FPWHFITDKR LELVQLGAGF MRLFGTHLAT HGS SLGTYF RLLRPRGVPL DFREILKRVN TPFMFCLKMP GSTALAEGLE IKGQMVFCAE SDSLLFVGSP FLDGLEGLTG RGLF ISDIP LHDATRDVIL VGEQARAQDG LRRRMDKLKN SIEEASKAVD KEREKNVSLL HLIFPPHIAK RLWLGEKIEA KSHDD VTML FSDIVGFTSI CATATPMMVI AMLEDLYSVF DIFCEELDVY KVETIGDAYC VASGLHRKVE THAPQIAWMA LRMVET CAQ HLTHEGNPIK MRIGLHTGTV LAGVVGKTML KYCLFGHNVT LANKFESGSE PLKINVSPTT YEWLIKFPGF DMEPRDR SC LPNSFPKDIH GTCYFLHKYT HPGTDPGEPQ VKHIREALKD YGIGQANSTD VDTEEPT UniProtKB: guanylate cyclase |
-Macromolecule #2: Soluble guanylyl cyclase beta-1 subunit
| Macromolecule | Name: Soluble guanylyl cyclase beta-1 subunit / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Manduca sexta (tobacco hornworm) Manduca sexta (tobacco hornworm) |
| Molecular weight | Theoretical: 68.182 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 2 RZ-2014 (butterflies/moths) Spodoptera aff. frugiperda 2 RZ-2014 (butterflies/moths) |
| Sequence | String: MYGFVNYALE LLVMKTFDEE TWETIKKKAD VAMEGSFLVR QIYEDEITYN LITAAVEVLQ IPADAILELF GKTFFEFCQD SGYDKILQV LGATPRDFLQ NLDGLHDHLG TLYPGMRSPS FRCTERPEDG ALVLHYYSDR PGLEHIVIGI VKTVASKLHN T EVKVEILK ...String: MYGFVNYALE LLVMKTFDEE TWETIKKKAD VAMEGSFLVR QIYEDEITYN LITAAVEVLQ IPADAILELF GKTFFEFCQD SGYDKILQV LGATPRDFLQ NLDGLHDHLG TLYPGMRSPS FRCTERPEDG ALVLHYYSDR PGLEHIVIGI VKTVASKLHN T EVKVEILK TKEECDHVQF LITETSTTGR VSAPEIAEIE TLSLEPKVSP ATFCRVFPFH LMFDRDLNIV QAGRTVSRLL PR VTRPGCK ITDVLDTVRP HLEMTFANVL AHINTVYVLK TKPEEMSVTD PHEEIASLRL KGQMLYIPET DVVVFQCYPS VTN LDDLTR RGLCIADIPL HDATRDLVLM SEQFEADYKL TQNLEVLTDK LQQTFRELEL EKQKTDRLLY SVLPISVATE LRHR RPVPA RRYDTVTLLF SGIVGFANYC ARNSDHKGAM KIVRMLNDLY TAFDVLTDPK RNPNVYKVET VGDKYMAVSG LPEYE VAHA KHISLLALDM MDLSQTVTVD GEPVGITIGI HSGEVVTGVI GHRMPRYCLF GNTVNLTSRC ETTGVPGTIN VSEDTY NYL MREDNHDEQF ELTYRGHVTM KGKAEPMQTW FLTRKIH UniProtKB: Guanylate cyclase soluble subunit beta-1 |
-Macromolecule #3: PROTOPORPHYRIN IX CONTAINING FE
| Macromolecule | Name: PROTOPORPHYRIN IX CONTAINING FE / type: ligand / ID: 3 / Number of copies: 1 / Formula: HEM |
|---|---|
| Molecular weight | Theoretical: 616.487 Da |
| Chemical component information |  ChemComp-HEM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)