[English] 日本語
 Yorodumi
Yorodumi- EMDB-19994: Non-uniform refinement of the CryoEM structure of DeCLIC nanodisc... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Non-uniform refinement of the CryoEM structure of DeCLIC nanodisc with 10mM calcium | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ion channel / TRANSLOCASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationextracellular ligand-gated monoatomic ion channel activity / transmembrane signaling receptor activity / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Desulfofustis sp. PB-SRB1 (bacteria) Desulfofustis sp. PB-SRB1 (bacteria) | |||||||||
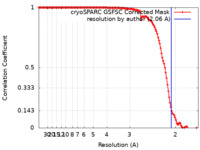
| Method | single particle reconstruction / cryo EM / Resolution: 2.06 Å | |||||||||
 Authors Authors | Fan C / Howard RJ / Lindahl E | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Calcium stabilization of a flexible N-terminal domain in a pentameric ligand-gated ion channel Authors: Fan C / Howard RJ / Lindahl E | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19994.map.gz emd_19994.map.gz | 197.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19994-v30.xml emd-19994-v30.xml emd-19994.xml emd-19994.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19994_fsc.xml emd_19994_fsc.xml | 12.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_19994.png emd_19994.png | 191.1 KB | ||
| Filedesc metadata |  emd-19994.cif.gz emd-19994.cif.gz | 6 KB | ||
| Others |  emd_19994_half_map_1.map.gz emd_19994_half_map_1.map.gz emd_19994_half_map_2.map.gz emd_19994_half_map_2.map.gz | 194.5 MB 194.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19994 http://ftp.pdbj.org/pub/emdb/structures/EMD-19994 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19994 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19994 | HTTPS FTP |
-Validation report
| Summary document |  emd_19994_validation.pdf.gz emd_19994_validation.pdf.gz | 765.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19994_full_validation.pdf.gz emd_19994_full_validation.pdf.gz | 765 KB | Display | |
| Data in XML |  emd_19994_validation.xml.gz emd_19994_validation.xml.gz | 21.3 KB | Display | |
| Data in CIF |  emd_19994_validation.cif.gz emd_19994_validation.cif.gz | 27.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19994 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19994 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19994 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19994 | HTTPS FTP |
-Related structure data
| Related structure data |  9ev8MC  9ev1C  9ev7C  9ev9C  9evaC  9evbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19994.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19994.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8464 Å | ||||||||||||||||||||||||||||||||||||
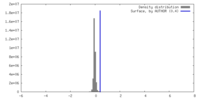
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_19994_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
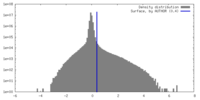
| Density Histograms |
-Half map: #1
| File | emd_19994_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
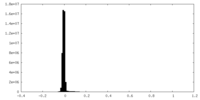
| Density Histograms |
- Sample components
Sample components
-Entire : DeCLIC in 10mM calcium
| Entire | Name: DeCLIC in 10mM calcium |
|---|---|
| Components |
|
-Supramolecule #1: DeCLIC in 10mM calcium
| Supramolecule | Name: DeCLIC in 10mM calcium / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Desulfofustis sp. PB-SRB1 (bacteria) Desulfofustis sp. PB-SRB1 (bacteria) |
-Macromolecule #1: Neur_chan_LBD domain-containing protein
| Macromolecule | Name: Neur_chan_LBD domain-containing protein / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Desulfofustis sp. PB-SRB1 (bacteria) Desulfofustis sp. PB-SRB1 (bacteria) |
| Molecular weight | Theoretical: 71.733992 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHNLQQLLPT RSLIWIFSFL TSISIWCTVA HAETEGRVQH FTGYIEDGRG IFYSLPDMKQ GDIIYASMQN TGGNLDPLVG IMAEEIDPA VSLGQVLEKA LASENDLISE LTAVADRIFL GWDDDGGKGY SASLEFTIPR DGTYHIFAGS TITNQRLDKF Q PTYTTGSF ...String: MHNLQQLLPT RSLIWIFSFL TSISIWCTVA HAETEGRVQH FTGYIEDGRG IFYSLPDMKQ GDIIYASMQN TGGNLDPLVG IMAEEIDPA VSLGQVLEKA LASENDLISE LTAVADRIFL GWDDDGGKGY SASLEFTIPR DGTYHIFAGS TITNQRLDKF Q PTYTTGSF QLILGLNAPQ VISGEGEPEG EVFASLASLE IKPEAHVQEL EIRLDKDTRY LTQHTRNLQP GDTFHALVEP IG EAPLPRL RLTDSGGKPL AFGLIDQPGE SVELNYTCDQ DICELVVHVD GTDGQKDSGE AVYRLLVGIN APNLRESGQT PVG SSVFLE SDLVTVGLAV DQIVGVDQRS ENFSVVGTLK LSWHDPKLGF SPDQCGCTVK SFEDASIRAV AGEINLPLPS FSFY NQQGN RWSQNQVIFV TPDGRASYFE RFTVTLQAPD FDFLAYPFDR QKFSIKVDLA VPTNMFIFNE IERFQQVVGD QLGEE EWVV TSYSQEITEV PFERGSTNSR FTTTLLVKRN LEYYILRIFV PLFLIISVSW VIFFLKDYGR QLEVASGNLL VFVAFN FTI SGDLPRLGYL TVLDRFMIVS FCLTAIVVLI SVCQKRLGAV GKQAVAAQID TWVLVIYPLV YSLYIIWVYL RFFTDHI GW UniProtKB: Uncharacterized protein |
-Macromolecule #2: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 2 / Number of copies: 10 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #3: N-OCTANE
| Macromolecule | Name: N-OCTANE / type: ligand / ID: 3 / Number of copies: 10 / Formula: OCT |
|---|---|
| Molecular weight | Theoretical: 114.229 Da |
| Chemical component information |  ChemComp-OCT: |
-Macromolecule #4: DODECANE
| Macromolecule | Name: DODECANE / type: ligand / ID: 4 / Number of copies: 10 / Formula: D12 |
|---|---|
| Molecular weight | Theoretical: 170.335 Da |
| Chemical component information |  ChemComp-D12: |
-Macromolecule #5: DECANE
| Macromolecule | Name: DECANE / type: ligand / ID: 5 / Number of copies: 5 / Formula: D10 |
|---|---|
| Molecular weight | Theoretical: 142.282 Da |
| Chemical component information |  ChemComp-D10: |
-Macromolecule #6: TETRADECANE
| Macromolecule | Name: TETRADECANE / type: ligand / ID: 6 / Number of copies: 5 / Formula: C14 |
|---|---|
| Molecular weight | Theoretical: 198.388 Da |
| Chemical component information |  ChemComp-C14: |
-Macromolecule #7: 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE
| Macromolecule | Name: 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE / type: ligand / ID: 7 / Number of copies: 5 / Formula: PX6 |
|---|---|
| Molecular weight | Theoretical: 647.883 Da |
| Chemical component information |  ChemComp-PX6: |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 215 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)