[English] 日本語
 Yorodumi
Yorodumi- EMDB-19121: CryoEM structure of the plant helper NLR NRC2 in its resting state -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of the plant helper NLR NRC2 in its resting state | |||||||||
 Map data Map data | Please use contour for 0.0034 for atom inclusion. This covers all the main chain atoms in the model | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NLR protein / Helper NLRl / plant immunity / R-protein / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationdefense response to other organism / ADP binding / defense response to virus / ATP binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
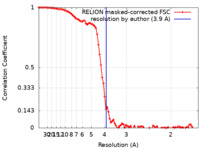
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Selvaraj M / Kamoun S / Contreras MP | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Biol / Year: 2024 Journal: PLoS Biol / Year: 2024Title: Activation of plant immunity through conversion of a helper NLR homodimer into a resistosome. Authors: Muniyandi Selvaraj / AmirAli Toghani / Hsuan Pai / Yu Sugihara / Jiorgos Kourelis / Enoch Lok Him Yuen / Tarhan Ibrahim / He Zhao / Rongrong Xie / Abbas Maqbool / Juan Carlos De la ...Authors: Muniyandi Selvaraj / AmirAli Toghani / Hsuan Pai / Yu Sugihara / Jiorgos Kourelis / Enoch Lok Him Yuen / Tarhan Ibrahim / He Zhao / Rongrong Xie / Abbas Maqbool / Juan Carlos De la Concepcion / Mark J Banfield / Lida Derevnina / Benjamin Petre / David M Lawson / Tolga O Bozkurt / Chih-Hang Wu / Sophien Kamoun / Mauricio P Contreras /  Abstract: Nucleotide-binding domain and leucine-rich repeat (NLR) proteins can engage in complex interactions to detect pathogens and execute a robust immune response via downstream helper NLRs. However, the ...Nucleotide-binding domain and leucine-rich repeat (NLR) proteins can engage in complex interactions to detect pathogens and execute a robust immune response via downstream helper NLRs. However, the biochemical mechanisms of helper NLR activation by upstream sensor NLRs remain poorly understood. Here, we show that the coiled-coil helper NLR NRC2 from Nicotiana benthamiana accumulates in vivo as a homodimer that converts into a higher-order oligomer upon activation by its upstream virus disease resistance protein Rx. The cryo-EM structure of NbNRC2 in its resting state revealed intermolecular interactions that mediate homodimer formation and contribute to immune receptor autoinhibition. These dimerization interfaces have diverged between paralogous NRC proteins to insulate critical network nodes and enable redundant immune pathways, possibly to minimise undesired cross-activation and evade pathogen suppression of immunity. Our results expand the molecular mechanisms of NLR activation pointing to transition from homodimers to higher-order oligomeric resistosomes. #1:  Journal: Biorxiv / Year: 2023 Journal: Biorxiv / Year: 2023Title: Activation of plant immunity through conversion of a helper NLR homodimer into a resistosome Authors: Selvaraj M / Kamoun S / Contreras MP | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19121.map.gz emd_19121.map.gz | 8.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19121-v30.xml emd-19121-v30.xml emd-19121.xml emd-19121.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19121_fsc.xml emd_19121_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_19121.png emd_19121.png | 57.1 KB | ||
| Masks |  emd_19121_msk_1.map emd_19121_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19121.cif.gz emd-19121.cif.gz | 7.3 KB | ||
| Others |  emd_19121_half_map_1.map.gz emd_19121_half_map_1.map.gz emd_19121_half_map_2.map.gz emd_19121_half_map_2.map.gz | 48.5 MB 48.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19121 http://ftp.pdbj.org/pub/emdb/structures/EMD-19121 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19121 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19121 | HTTPS FTP |
-Related structure data
| Related structure data |  8rfhMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19121.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19121.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Please use contour for 0.0034 for atom inclusion. This covers all the main chain atoms in the model | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.828 Å | ||||||||||||||||||||||||||||||||||||
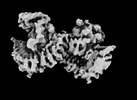
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19121_msk_1.map emd_19121_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_19121_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_19121_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : NRC2 heler NLR dimer with ADP
| Entire | Name: NRC2 heler NLR dimer with ADP |
|---|---|
| Components |
|
-Supramolecule #1: NRC2 heler NLR dimer with ADP
| Supramolecule | Name: NRC2 heler NLR dimer with ADP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: This is helper NLR protein NRC2 in inactive state with ADP nucleotide bound state |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 100 KDa |
-Macromolecule #1: NRC2a
| Macromolecule | Name: NRC2a / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 107.522398 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MANVAVEFLV QNLMQLLRDN AELIVGVKDS AESLLQDLND FNAFLKQTAK SRTENDVHKE LVKKIKTVVN SAEDAIDKFV IEAKLHKDK GVGRFVDVKH YKRVYDVAGE IKTIRDKVKE IRLNNALDLQ ALQDEDQSAK GVQERKPPVV EEDDVVGFEE E ADKVINRL ...String: MANVAVEFLV QNLMQLLRDN AELIVGVKDS AESLLQDLND FNAFLKQTAK SRTENDVHKE LVKKIKTVVN SAEDAIDKFV IEAKLHKDK GVGRFVDVKH YKRVYDVAGE IKTIRDKVKE IRLNNALDLQ ALQDEDQSAK GVQERKPPVV EEDDVVGFEE E ADKVINRL LGGSSGLEVV PVVGMPGLGK TTLANKIYKH PDIGYQFFTR IWVYVSQSYR RRELFLNIIS KFTRNTKQYH DM CEEDLAD EIEDFLGKGG KYLIVLDDVW SPDAWERIRI AFPNNNKSNR ILLTTRDSKV AKQCKQCIGI PHDLKFLTED ESW ILLEKK VFHKDKCPPE LELSGKSIAK KCNGLPLAIV VIAGALIGKG KTSREWKQVD ESVGEHLINK DQPENCNKLV QLSY DRLSY DLKACFLYCG AFPGGFEIPA WKLIRLWIAE GFIQYKGHLS LECKAEDNLN DLINRNLVMV MQRTSDGQIK TCRLH DMLH EFCRQEAMKE ENLFQEIKLG AEQYFPGKRE LATYRRLCIH SSVLEFISTK PSGEHVRSFL SFSLKKIEMP SVDIPT IPK GFPLLRVFDV ESINFSRFSK EFFQLYHLRY IAFSSDTIKI IPKHIGELWN IQTLIINTQQ RSLDIQANIW NMERLRH LH TNSSAKLPVP VTPRSSKVPL VNQSLQTLST IAPESCTEEV FARTPNLKKL GIRGKIAVLL EPNKSLLKNV KKLESLEN L KLINDSSQTG KGLRLPPSYI FPTKLRKLSL VDTWLEWNDM SILGQMEHLE VLKLKENGFM GECWESVGGF CSLLVLWIE RTDLVSWKAS ADHFPRLKHL VLICCDKLKE IPIGLADIRS FQVMELQNST KTAAISARGI RDKKDKQTQE GTNNNGFKLS IFPPDLGSG SGRGSHHHHH HDYDIPTTAS ENLYFQGELD YKDHDGDYKD HDIDYKDDDD K UniProtKB: NRC2a |
-Macromolecule #2: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: C-flat / Material: COPPER / Support film - Material: GRAPHENE / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Details: The grids were negatively glow discharged | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: The C-flat grids were coated with grphene oxide, in house and the sample was applied two times inside the vitrobot chamber. | ||||||||||||
| Details | the particles are NRC2 purified from plant after over expression. This was suspended in buffer and cryogrids were made on graphene oxide backing |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 6000 / Average exposure time: 2.6 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Residue range: 1-880 / Chain - Source name: AlphaFold / Chain - Initial model type: in silico model / Details: This is the full length sequence of NRC |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 100 / Target criteria: CC |
| Output model |  PDB-8rfh: |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)