[English] 日本語
 Yorodumi
Yorodumi- EMDB-19009: Cryo-EM structure of the inward-facing ethanolamine-bound FLVCR1 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the inward-facing ethanolamine-bound FLVCR1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | MFS / ethanolamine transport / human transporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationheme export / ethanolamine transmembrane transporter activity / heme transport / choline transmembrane transporter activity / heme transmembrane transporter activity / embryonic skeletal system morphogenesis / choline transport / phospholipid biosynthetic process / head morphogenesis / regulation of organ growth ...heme export / ethanolamine transmembrane transporter activity / heme transport / choline transmembrane transporter activity / heme transmembrane transporter activity / embryonic skeletal system morphogenesis / choline transport / phospholipid biosynthetic process / head morphogenesis / regulation of organ growth / Heme biosynthesis / heme biosynthetic process / embryonic digit morphogenesis / erythrocyte maturation / blood vessel development / mitochondrial transport / spleen development / erythrocyte differentiation / Iron uptake and transport / multicellular organism growth / in utero embryonic development / intracellular iron ion homeostasis / mitochondrial inner membrane / heme binding / mitochondrion / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Weng T-H / Wu D / Safarian S | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Molecular mechanism of choline and ethanolamine transport in humans. Authors: Keiken Ri / Tsai-Hsuan Weng / Ainara Claveras Cabezudo / Wiebke Jösting / Yu Zhang / Andre Bazzone / Nancy C P Leong / Sonja Welsch / Raymond T Doty / Gonca Gursu / Tiffany Jia Ying Lim / ...Authors: Keiken Ri / Tsai-Hsuan Weng / Ainara Claveras Cabezudo / Wiebke Jösting / Yu Zhang / Andre Bazzone / Nancy C P Leong / Sonja Welsch / Raymond T Doty / Gonca Gursu / Tiffany Jia Ying Lim / Sarah Luise Schmidt / Janis L Abkowitz / Gerhard Hummer / Di Wu / Long N Nguyen / Schara Safarian /    Abstract: Human feline leukaemia virus subgroup C receptor-related proteins 1 and 2 (FLVCR1 and FLVCR2) are members of the major facilitator superfamily. Their dysfunction is linked to several clinical ...Human feline leukaemia virus subgroup C receptor-related proteins 1 and 2 (FLVCR1 and FLVCR2) are members of the major facilitator superfamily. Their dysfunction is linked to several clinical disorders, including PCARP, HSAN and Fowler syndrome. Earlier studies concluded that FLVCR1 may function as a haem exporter, whereas FLVCR2 was suggested to act as a haem importer, yet conclusive biochemical and detailed molecular evidence remained elusive for the function of both transporters. Here, we show that FLVCR1 and FLVCR2 facilitate the transport of choline and ethanolamine across the plasma membrane, using a concentration-driven substrate translocation process. Through structural and computational analyses, we have identified distinct conformational states of FLVCRs and unravelled the coordination chemistry underlying their substrate interactions. Fully conserved tryptophan and tyrosine residues form the binding pocket of both transporters and confer selectivity for choline and ethanolamine through cation-π interactions. Our findings clarify the mechanisms of choline and ethanolamine transport by FLVCR1 and FLVCR2, enhance our comprehension of disease-associated mutations that interfere with these vital processes and shed light on the conformational dynamics of these major facilitator superfamily proteins during the transport cycle. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19009.map.gz emd_19009.map.gz | 12 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19009-v30.xml emd-19009-v30.xml emd-19009.xml emd-19009.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19009_fsc.xml emd_19009_fsc.xml | 5.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_19009.png emd_19009.png | 117.1 KB | ||
| Filedesc metadata |  emd-19009.cif.gz emd-19009.cif.gz | 6.2 KB | ||
| Others |  emd_19009_half_map_1.map.gz emd_19009_half_map_1.map.gz emd_19009_half_map_2.map.gz emd_19009_half_map_2.map.gz | 9.8 MB 9.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19009 http://ftp.pdbj.org/pub/emdb/structures/EMD-19009 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19009 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19009 | HTTPS FTP |
-Related structure data
| Related structure data |  8r8tMC  8qcsC  8qctC  8qcxC  8qcyC  8qd0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19009.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19009.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.146 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_19009_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
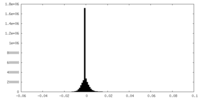
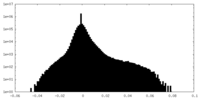
| Density Histograms |
-Half map: #2
| File | emd_19009_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
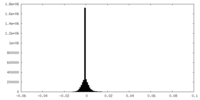
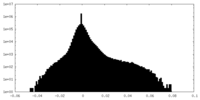
| Density Histograms |
- Sample components
Sample components
-Entire : ethanolamine-bound FLVCR1 monomer
| Entire | Name: ethanolamine-bound FLVCR1 monomer |
|---|---|
| Components |
|
-Supramolecule #1: ethanolamine-bound FLVCR1 monomer
| Supramolecule | Name: ethanolamine-bound FLVCR1 monomer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 60 KDa |
-Macromolecule #1: Heme transporter FLVCR1
| Macromolecule | Name: Heme transporter FLVCR1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 60.896316 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MARPDDEEGA AVAPGHPLAK GYLPLPRGAP VGKESVELQN GPKAGTFPVN GAPRDSLAAA SGVLGGPQTP LAPEEETQAR LLPAGAGAE TPGAESSPLP LTALSPRRFV VLLIFSLYSL VNAFQWIQYS IISNVFEGFY GVTLLHIDWL SMVYMLAYVP L IFPATWLL ...String: MARPDDEEGA AVAPGHPLAK GYLPLPRGAP VGKESVELQN GPKAGTFPVN GAPRDSLAAA SGVLGGPQTP LAPEEETQAR LLPAGAGAE TPGAESSPLP LTALSPRRFV VLLIFSLYSL VNAFQWIQYS IISNVFEGFY GVTLLHIDWL SMVYMLAYVP L IFPATWLL DTRGLRLTAL LGSGLNCLGA WIKCGSVQQH LFWVTMLGQC LCSVAQVFIL GLPSRIASVW FGPKEVSTAC AT AVLGNQL GTAVGFLLPP VLVPNTQNDT NLLACNISTM FYGTSAVATL LFILTAIAFK EKPRYPPSQA QAALQDSPPE EYS YKKSIR NLFKNIPFVL LLITYGIMTG AFYSVSTLLN QMILTYYEGE EVNAGRIGLT LVVAGMVGSI LCGLWLDYTK TYKQ TTLIV YILSFIGMVI FTFTLDLRYI IIVFVTGGVL GFFMTGYLPL GFEFAVEITY PESEGTSSGL LNASAQIFGI LFTLA QGKL TSDYGPKAGN IFLCVWMFIG IILTALIKSD LRRHNINIGI TNVDVKAIPA DSPTDQEPKT VMLSKQSESA IDYKDD DDK UniProtKB: Choline/ethanolamine transporter FLVCR1 |
-Macromolecule #2: ETHANOLAMINE
| Macromolecule | Name: ETHANOLAMINE / type: ligand / ID: 2 / Number of copies: 1 / Formula: ETA |
|---|---|
| Molecular weight | Theoretical: 61.083 Da |
| Chemical component information | 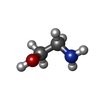 ChemComp-ETA: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.1 µm / Nominal defocus min: 1.1 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)