[English] 日本語
 Yorodumi
Yorodumi- EMDB-1858: Structure of the ribosome-SecYE complex in the membrane environment -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1858 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the ribosome-SecYE complex in the membrane environment | |||||||||
 Map data Map data | This map represents an E.coli 70S ribosome carrying an elongation arrested nascent chain of 118 amino acid residues, with the first 102 residues representing the N-terminus of the membrane protein FtsQ with the signal anchor, a tRNA in the P-site and the E.coli SecYEG complex embedded in a E.coli lipid bilayer (Nanodisc). | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationDefective ABCA1 causes TGD / high-density lipoprotein particle receptor binding / peptidyl-methionine modification / HDL clearance / cell septum assembly / spherical high-density lipoprotein particle / Scavenging by Class B Receptors / negative regulation of response to cytokine stimulus / protein oxidation / regulation of intestinal cholesterol absorption ...Defective ABCA1 causes TGD / high-density lipoprotein particle receptor binding / peptidyl-methionine modification / HDL clearance / cell septum assembly / spherical high-density lipoprotein particle / Scavenging by Class B Receptors / negative regulation of response to cytokine stimulus / protein oxidation / regulation of intestinal cholesterol absorption / vitamin transport / blood vessel endothelial cell migration / cholesterol import / negative regulation of heterotypic cell-cell adhesion / apolipoprotein receptor binding / apolipoprotein A-I receptor binding / ABC transporters in lipid homeostasis / negative regulation of cell adhesion molecule production / protein transport by the Sec complex / negative regulation of cytokine production involved in immune response / intracellular protein transmembrane transport / HDL assembly / high-density lipoprotein particle binding / negative regulation of very-low-density lipoprotein particle remodeling / phosphatidylcholine biosynthetic process / glucocorticoid metabolic process / protein-transporting ATPase activity / acylglycerol homeostasis / phosphatidylcholine-sterol O-acyltransferase activator activity / positive regulation of phospholipid efflux / Chylomicron remodeling / cellular response to lipoprotein particle stimulus / Chylomicron assembly / high-density lipoprotein particle clearance / phospholipid efflux / chylomicron / high-density lipoprotein particle remodeling / positive regulation of cholesterol metabolic process / reverse cholesterol transport / lipid storage / phospholipid homeostasis / high-density lipoprotein particle assembly / chemorepellent activity / low-density lipoprotein particle / lipoprotein biosynthetic process / cholesterol transfer activity / cholesterol transport / high-density lipoprotein particle / very-low-density lipoprotein particle / FtsZ-dependent cytokinesis / endothelial cell proliferation / regulation of Cdc42 protein signal transduction / HDL remodeling / cholesterol efflux / Scavenging by Class A Receptors / triglyceride homeostasis / adrenal gland development / negative chemotaxis / negative regulation of interleukin-1 beta production / cholesterol binding / cell division site / cholesterol biosynthetic process / amyloid-beta formation / stringent response / positive regulation of Rho protein signal transduction / ornithine decarboxylase inhibitor activity / transcription antitermination factor activity, RNA binding / protein secretion / misfolded RNA binding / Group I intron splicing / protein transmembrane transporter activity / endocytic vesicle / RNA folding / positive regulation of cholesterol efflux / membrane => GO:0016020 / negative regulation of tumor necrosis factor-mediated signaling pathway / transcriptional attenuation / Scavenging of heme from plasma / endoribonuclease inhibitor activity / positive regulation of ribosome biogenesis / RNA-binding transcription regulator activity / protein targeting / Retinoid metabolism and transport / cholesterol metabolic process / translational termination / negative regulation of cytoplasmic translation / four-way junction DNA binding / positive regulation of stress fiber assembly / DnaA-L2 complex / heat shock protein binding / translation repressor activity / negative regulation of translational initiation / endocytic vesicle lumen / regulation of mRNA stability / negative regulation of DNA-templated DNA replication initiation / positive regulation of substrate adhesion-dependent cell spreading / mRNA regulatory element binding translation repressor activity / positive regulation of phagocytosis / cholesterol homeostasis / positive regulation of RNA splicing Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.1 Å | |||||||||
 Authors Authors | Frauenfeld J / Gumbart J / Sluis EO / Funes S / Gartmann M / Beatrix B / Mielke T / Berninghausen O / Becker T / Schulten K / Beckmann R | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2011 Journal: Nat Struct Mol Biol / Year: 2011Title: Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Authors: Jens Frauenfeld / James Gumbart / Eli O van der Sluis / Soledad Funes / Marco Gartmann / Birgitta Beatrix / Thorsten Mielke / Otto Berninghausen / Thomas Becker / Klaus Schulten / Roland Beckmann /  Abstract: The ubiquitous SecY-Sec61 complex translocates nascent secretory proteins across cellular membranes and integrates membrane proteins into lipid bilayers. Several structures of mostly detergent- ...The ubiquitous SecY-Sec61 complex translocates nascent secretory proteins across cellular membranes and integrates membrane proteins into lipid bilayers. Several structures of mostly detergent-solubilized Sec complexes have been reported. Here we present a single-particle cryo-EM structure of the SecYEG complex in a membrane environment, bound to a translating ribosome, at subnanometer resolution. Using the SecYEG complex reconstituted in a so-called Nanodisc, we could trace the nascent polypeptide chain from the peptidyltransferase center into the membrane. The reconstruction allowed for the identification of ribosome-lipid interactions. The rRNA helix 59 (H59) directly contacts the lipid surface and appears to modulate the membrane in immediate vicinity to the proposed lateral gate of the protein-conducting channel (PCC). On the basis of our map and molecular dynamics simulations, we present a model of a signal anchor-gated PCC in the membrane. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1858.map.gz emd_1858.map.gz | 8.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1858-v30.xml emd-1858-v30.xml emd-1858.xml emd-1858.xml | 9.9 KB 9.9 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-1858_image.jpg EMD-1858_image.jpg | 322.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1858 http://ftp.pdbj.org/pub/emdb/structures/EMD-1858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1858 | HTTPS FTP |
-Related structure data
| Related structure data |  4v6mMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1858.map.gz / Format: CCP4 / Size: 122.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1858.map.gz / Format: CCP4 / Size: 122.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This map represents an E.coli 70S ribosome carrying an elongation arrested nascent chain of 118 amino acid residues, with the first 102 residues representing the N-terminus of the membrane protein FtsQ with the signal anchor, a tRNA in the P-site and the E.coli SecYEG complex embedded in a E.coli lipid bilayer (Nanodisc). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2375 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
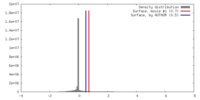
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : An active E.coli SecYEG complex embedded in a lipid bilayer (Nano...
| Entire | Name: An active E.coli SecYEG complex embedded in a lipid bilayer (Nanodisc), bound to a translating E.coli ribosome |
|---|---|
| Components |
|
-Supramolecule #1000: An active E.coli SecYEG complex embedded in a lipid bilayer (Nano...
| Supramolecule | Name: An active E.coli SecYEG complex embedded in a lipid bilayer (Nanodisc), bound to a translating E.coli ribosome type: sample / ID: 1000 Details: The heterotrimeric SecYEG complex was embedded in a lipid bilayer (nascent HDL, Nanodisc) Oligomeric state: 70S ribosome bound to one copy of the E.coli SecYEG complex in a lipid bilayer Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 2.7 MDa |
-Supramolecule #1: 70S ribosome nascent chain complex
| Supramolecule | Name: 70S ribosome nascent chain complex / type: complex / ID: 1 / Name.synonym: 70S RNC / Recombinant expression: No / Ribosome-details: ribosome-prokaryote: ALL |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.7 MDa |
-Macromolecule #1: Nd-SecYEG
| Macromolecule | Name: Nd-SecYEG / type: ligand / ID: 1 / Name.synonym: SecYEG embedded in a Nanodisc / Number of copies: 1 / Oligomeric state: Heterotrimeric / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 Details: 20 mM Hepes (pH 7.2), 100 mM KOAc, 10 mM Mg(OAc)2, 1 mM DTT, 250 microg/ml chloramphenicol |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: PRIMESCAN / Digitization - Sampling interval: 4.76 µm / Average electron dose: 22 e/Å2 / Details: Scanned at 5334 dpi |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 38000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.26 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 39000 |
| Sample stage | Specimen holder: FEI Polara cartridge system / Specimen holder model: OTHER |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Defocus group volumes |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 7.1 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER / Number images used: 85664 |
 Movie
Movie Controller
Controller































 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)