+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Ammonium Transporter Amt1 from Shewanella denitrificans | |||||||||
 Map data Map data | unsharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ammonium Transporter Amt1 / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationammonium homeostasis / ammonium channel activity / catalytic activity / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Shewanella denitrificans (bacteria) Shewanella denitrificans (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.53 Å | |||||||||
 Authors Authors | Gschell M / Zhang L / Einsle O / Andrade S | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: How sensor Amt-like proteins integrate ammonium signals. Authors: Tobias Pflüger / Mathias Gschell / Lin Zhang / Volodymyr Shnitsar / Annas J Zabadné / Paul Zierep / Stefan Günther / Oliver Einsle / Susana L A Andrade /  Abstract: Unlike aquaporins or potassium channels, ammonium transporters (Amts) uniquely discriminate ammonium from potassium and water. This feature has certainly contributed to their repurposing as ammonium ...Unlike aquaporins or potassium channels, ammonium transporters (Amts) uniquely discriminate ammonium from potassium and water. This feature has certainly contributed to their repurposing as ammonium receptors during evolution. Here, we describe the ammonium receptor Sd-Amt1, where an Amt module connects to a cytoplasmic diguanylate cyclase transducer module via an HAMP domain. Structures of the protein with and without bound ammonium were determined to 1.7- and 1.9-Ångstrom resolution, depicting the ON and OFF states of the receptor and confirming the presence of a binding site for two ammonium cations that is pivotal for signal perception and receptor activation. The transducer domain was disordered in the crystals, and an AlphaFold2 prediction suggests that the helices linking both domains are flexible. While the sensor domain retains the trimeric fold formed by all Amt family members, the HAMP domains interact as pairs and serve to dimerize the transducer domain upon activation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18549.map.gz emd_18549.map.gz | 27.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18549-v30.xml emd-18549-v30.xml emd-18549.xml emd-18549.xml | 16.5 KB 16.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_18549.png emd_18549.png | 126.8 KB | ||
| Filedesc metadata |  emd-18549.cif.gz emd-18549.cif.gz | 5.6 KB | ||
| Others |  emd_18549_additional_1.map.gz emd_18549_additional_1.map.gz emd_18549_half_map_1.map.gz emd_18549_half_map_1.map.gz emd_18549_half_map_2.map.gz emd_18549_half_map_2.map.gz | 28.3 MB 28.3 MB 28.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18549 http://ftp.pdbj.org/pub/emdb/structures/EMD-18549 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18549 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18549 | HTTPS FTP |
-Related structure data
| Related structure data |  8qpfMC  8qj3C  8qj4C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18549.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18549.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
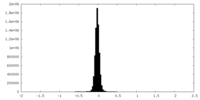
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: sharpened map
| File | emd_18549_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
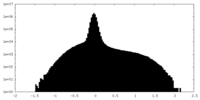
| Density Histograms |
-Half map: half map A
| File | emd_18549_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
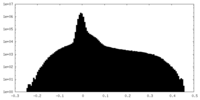
| Density Histograms |
-Half map: half map A
| File | emd_18549_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : homotrimer Amt1
| Entire | Name: homotrimer Amt1 |
|---|---|
| Components |
|
-Supramolecule #1: homotrimer Amt1
| Supramolecule | Name: homotrimer Amt1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Shewanella denitrificans (bacteria) Shewanella denitrificans (bacteria) |
-Macromolecule #1: Ammonium transporter
| Macromolecule | Name: Ammonium transporter / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Shewanella denitrificans (bacteria) Shewanella denitrificans (bacteria) |
| Molecular weight | Theoretical: 73.021875 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSESFLLNLW ILLCACLVLI MQAGFTCFES GNVRNKNSVN VALKNVSDFC VCAVCYWAFG YALMYGNSID GIVGANGFFY STTTNSHET SFFLFQLMFC CTSATIISGA VAERMRFTGY ILVTLLAASL IYPLFGHWAW GGRILGSETS TPGWLEQLGF I DFAGATVV ...String: MSESFLLNLW ILLCACLVLI MQAGFTCFES GNVRNKNSVN VALKNVSDFC VCAVCYWAFG YALMYGNSID GIVGANGFFY STTTNSHET SFFLFQLMFC CTSATIISGA VAERMRFTGY ILVTLLAASL IYPLFGHWAW GGRILGSETS TPGWLEQLGF I DFAGATVV HSVGGWMALA CVLIIGPRLG RFNNKHGVNQ IFGDNLPLTA LGTFLLFLGW FGFNGGSYGK IDDMLSSVFV NT ALGGTFG GFVVLLICIW QQSLLSIRFV LNGVLAGLVA ITASANSISS IDAATIGGIS GALSFFATIL LEKCKIDDVV SVV PVHLIG GIWGTLALAI FADGQYFIAG NSRVDQFLIQ LLGVVTCGIF AFGLPYMLIR LLNRVYPLRV SPRVEILGLN FGEF GLKSE TFNFLKQMRK NKNSHKNKQA VSVDFFSDIG LIEAEYNAFL EVINLQQRQA DINSHRLSRL AKTDHLTRLN NRLGF DECY DSEWLRMRRE KKPFSLLLID IDHFKLYNDH YGHPRGDQCL QQVALVLAST AKRPADCCAR VGGEEFAILL PDTDSE AGE KIANDINIRL KALEIPHLSS PIMPYVTVSI GIATLTPERY DSLDQAYLYQ CADDALYSAK QAGRNGVKAV IIDEAHE QT KKLDENLYFQ GFEHHHHHH UniProtKB: Ammonium transporter |
-Macromolecule #2: water
| Macromolecule | Name: water / type: ligand / ID: 2 / Number of copies: 202 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: DARK FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.53 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 477507 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)