[English] 日本語
 Yorodumi
Yorodumi- EMDB-18352: S. cerevisia Niemann-Pick type C protein NCR1 in LMNG at pH 5.5 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | S. cerevisia Niemann-Pick type C protein NCR1 in LMNG at pH 5.5 | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | sterol transport / vacuole / lysosome / LIPID TRANSPORT / membrane protein | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationIntestinal lipid absorption / LDL clearance / sterol binding / sterol transport / sphingolipid metabolic process / fungal-type vacuole membrane / transmembrane transporter activity / endoplasmic reticulum / membrane Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
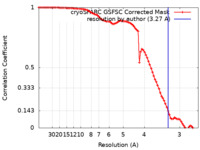
| Method | single particle reconstruction / cryo EM / Resolution: 3.27 Å | ||||||||||||
 Authors Authors | Frain KM / Nel L / Dedic E / Olesen E / Stokes D / Panyella Pedersen B | ||||||||||||
| Funding support |  United States, United States,  Denmark, 3 items Denmark, 3 items
| ||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Conformational changes in the Niemann-Pick type C1 protein NCR1 drive sterol translocation. Authors: Kelly M Frain / Emil Dedic / Lynette Nel / Anastasiia Bohush / Esben Olesen / Katja Thaysen / Daniel Wüstner / David L Stokes / Bjørn Panyella Pedersen /   Abstract: The membrane protein Niemann-Pick type C1 (NPC1, named NCR1 in yeast) is central to sterol homeostasis in eukaryotes. NCR1 is localized to the vacuolar membrane, where it is suggested to carry ...The membrane protein Niemann-Pick type C1 (NPC1, named NCR1 in yeast) is central to sterol homeostasis in eukaryotes. NCR1 is localized to the vacuolar membrane, where it is suggested to carry sterols across the protective glycocalyx and deposit them into the vacuolar membrane. However, documentation of a vacuolar glycocalyx in fungi is lacking, and the mechanism for sterol translocation has remained unclear. Here, we provide evidence supporting the presence of a glycocalyx in isolated vacuoles and report four cryo-EM structures of NCR1 in two distinct conformations, named tense and relaxed. These two conformations illustrate the movement of sterols through a tunnel formed by the luminal domains, thus bypassing the barrier presented by the glycocalyx. Based on these structures and on comparison with other members of the Resistance-Nodulation-Division (RND) superfamily, we propose a transport model that links changes in the luminal domains with a cycle of protonation and deprotonation within the transmembrane region of the protein. Our model suggests that NPC proteins work by a generalized RND mechanism where the proton motive force drives conformational changes in the transmembrane domains that are allosterically coupled to luminal/extracellular domains to promote sterol transport. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18352.map.gz emd_18352.map.gz | 56.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18352-v30.xml emd-18352-v30.xml emd-18352.xml emd-18352.xml | 21.8 KB 21.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18352_fsc.xml emd_18352_fsc.xml | 8.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_18352.png emd_18352.png | 31.1 KB | ||
| Masks |  emd_18352_msk_1.map emd_18352_msk_1.map | 59.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18352.cif.gz emd-18352.cif.gz | 7.6 KB | ||
| Others |  emd_18352_half_map_1.map.gz emd_18352_half_map_1.map.gz emd_18352_half_map_2.map.gz emd_18352_half_map_2.map.gz | 55.4 MB 55.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18352 http://ftp.pdbj.org/pub/emdb/structures/EMD-18352 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18352 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18352 | HTTPS FTP |
-Related structure data
| Related structure data |  8qedMC  8qebC  8qecC  8qeeC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18352.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18352.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.302 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18352_msk_1.map emd_18352_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_18352_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_18352_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : NCR1 pH 5.5 in LMNG
| Entire | Name: NCR1 pH 5.5 in LMNG |
|---|---|
| Components |
|
-Supramolecule #1: NCR1 pH 5.5 in LMNG
| Supramolecule | Name: NCR1 pH 5.5 in LMNG / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 132.6 KDa |
-Macromolecule #1: NPC intracellular sterol transporter 1-related protein 1
| Macromolecule | Name: NPC intracellular sterol transporter 1-related protein 1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 132.755094 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNVLWIIALV GQLMRLVQGT ATCAMYGNCG KKSVFGNELP CPVPRSFEPP VLSDETSKLL VEVCGEEWKE VRYACCTKDQ VVALRDNLQ KAQPLISSCP ACLKNFNNLF CHFTCAADQG RFVNITKVEK SKEDKDIVAE LDVFMNSSWA SEFYDSCKNI K FSATNGYA ...String: MNVLWIIALV GQLMRLVQGT ATCAMYGNCG KKSVFGNELP CPVPRSFEPP VLSDETSKLL VEVCGEEWKE VRYACCTKDQ VVALRDNLQ KAQPLISSCP ACLKNFNNLF CHFTCAADQG RFVNITKVEK SKEDKDIVAE LDVFMNSSWA SEFYDSCKNI K FSATNGYA MDLIGGGAKN YSQFLKFLGD AKPMLGGSPF QINYKYDLAN EEKEWQEFND EVYACDDAQY KCACSDCQES CP HLKPLKD GVCKVGPLPC FSLSVLIFYT ICALFAFMWY YLCKRKKNGA MIVDDDIVPE SGSLDESETN VFESFNNETN FFN GKLANL FTKVGQFSVE NPYKILITTV FSIFVFSFII FQYATLETDP INLWVSKNSE KFKEKEYFDD NFGPFYRTEQ IFVV NETGP VLSYETLHWW FDVENFITEE LQSSENIGYQ DLCFRPTEDS TCVIESFTQY FQGALPNKDS WKRELQECGK FPVNC LPTF QQPLKTNLLF SDDDILNAHA FVVTLLLTNH TQSANRWEER LEEYLLDLKV PEGLRISFNT EISLEKELNN NNDIST VAI SYLMMFLYAT WALRRKDGKT RLLLGISGLL IVLASIVCAA GFLTLFGLKS TLIIAEVIPF LILAIGIDNI FLITHEY DR NCEQKPEYSI DQKIISAIGR MSPSILMSLL CQTGCFLIAA FVTMPAVHNF AIYSTVSVIF NGVLQLTAYV SILSLYEK R SNYKQITGNE ETKESFLKTF YFKMLTQKRL IIIIFSAWFF TSLVFLPEIQ FGLDQTLAVP QDSYLVDYFK DVYSFLNVG PPVYMVVKNL DLTKRQNQQK ICGKFTTCER DSLANVLEQE RHRSTITEPL ANWLDDYFMF LNPQNDQCCR LKKGTDEVCP PSFPSRRCE TCFQQGSWNY NMSGFPEGKD FMEYLSIWIN APSDPCPLGG RAPYSTALVY NETSVSASVF RTAHHPLRSQ K DFIQAYSD GVRISSSFPE LDMFAYSPFY IFFVQYQTLG PLTLKLIGSA IILIFFISSV FLQNIRSSFL LALVVTMIIV DI GALMALL GISLNAVSLV NLIICVGLGV EFCVHIVRSF TVVPSETKKD ANSRVLYSLN TIGESVIKGI TLTKFIGVCV LAF AQSKIF DVFYFRMWFT LIIVAALHAL LFLPALLSLF GGESYRDDSI EAED UniProtKB: NPC intracellular sterol transporter 1-related protein 1 |
-Macromolecule #3: ERGOSTEROL
| Macromolecule | Name: ERGOSTEROL / type: ligand / ID: 3 / Number of copies: 2 / Formula: ERG |
|---|---|
| Molecular weight | Theoretical: 396.648 Da |
| Chemical component information |  ChemComp-ERG: |
-Macromolecule #4: PHOSPHATIDYLETHANOLAMINE
| Macromolecule | Name: PHOSPHATIDYLETHANOLAMINE / type: ligand / ID: 4 / Number of copies: 1 / Formula: PTY |
|---|---|
| Molecular weight | Theoretical: 734.039 Da |
| Chemical component information |  ChemComp-PTY: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 5.5 Component:
| ||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. | ||||||||||||
| Vitrification | Cryogen name: NITROGEN / Chamber humidity: 99 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5790 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 8541 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)