[English] 日本語
 Yorodumi
Yorodumi- EMDB-18308: Helical reconstruction of yeast eisosome protein Pil1 bound to me... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture -PIP2/+sterol (DOPC, DOPE, DOPS, cholesterol 30:20:20:30) | |||||||||||||||
 Map data Map data | Helical map Type II -PIP2/ sterol (D1, rise=5.04, twist=-136.50) | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | BAR domain / lipid reconstitution / membrane microdomain / LIPID BINDING PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationprotein localization to eisosome filament / eisosome filament / eisosome assembly / eisosome / lipid droplet / cell periphery / endocytosis / intracellular protein localization / mitochondrial outer membrane / lipid binding ...protein localization to eisosome filament / eisosome filament / eisosome assembly / eisosome / lipid droplet / cell periphery / endocytosis / intracellular protein localization / mitochondrial outer membrane / lipid binding / mitochondrion / plasma membrane / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.35 Å | |||||||||||||||
 Authors Authors | Kefauver JM / Zou L / Desfosses A / Loewith RJ | |||||||||||||||
| Funding support | European Union,  Switzerland, 4 items Switzerland, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain. Authors: Jennifer M Kefauver / Markku Hakala / Luoming Zou / Josephine Alba / Javier Espadas / Maria G Tettamanti / Jelena Gajić / Caroline Gabus / Pablo Campomanes / Leandro F Estrozi / Nesli E Sen ...Authors: Jennifer M Kefauver / Markku Hakala / Luoming Zou / Josephine Alba / Javier Espadas / Maria G Tettamanti / Jelena Gajić / Caroline Gabus / Pablo Campomanes / Leandro F Estrozi / Nesli E Sen / Stefano Vanni / Aurélien Roux / Ambroise Desfosses / Robbie Loewith /    Abstract: Biological membranes are partitioned into functional zones termed membrane microdomains, which contain specific lipids and proteins. The composition and organization of membrane microdomains remain ...Biological membranes are partitioned into functional zones termed membrane microdomains, which contain specific lipids and proteins. The composition and organization of membrane microdomains remain controversial because few techniques are available that allow the visualization of lipids in situ without disrupting their native behaviour. The yeast eisosome, composed of the BAR-domain proteins Pil1 and Lsp1 (hereafter, Pil1/Lsp1), scaffolds a membrane compartment that senses and responds to mechanical stress by flattening and releasing sequestered factors. Here we isolated near-native eisosomes as helical tubules made up of a lattice of Pil1/Lsp1 bound to plasma membrane lipids, and solved their structures by helical reconstruction. Our structures reveal a striking organization of membrane lipids, and, using in vitro reconstitutions and molecular dynamics simulations, we confirmed the positioning of individual PI(4,5)P, phosphatidylserine and sterol molecules sequestered beneath the Pil1/Lsp1 coat. Three-dimensional variability analysis of the native-source eisosomes revealed a dynamic stretching of the Pil1/Lsp1 lattice that affects the sequestration of these lipids. Collectively, our results support a mechanism in which stretching of the Pil1/Lsp1 lattice liberates lipids that would otherwise be anchored by the Pil1/Lsp1 coat, and thus provide mechanistic insight into how eisosome BAR-domain proteins create a mechanosensitive membrane microdomain. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18308.map.gz emd_18308.map.gz | 944.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18308-v30.xml emd-18308-v30.xml emd-18308.xml emd-18308.xml | 23.6 KB 23.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_18308.png emd_18308.png | 206.2 KB | ||
| Masks |  emd_18308_msk_1.map emd_18308_msk_1.map | 1000 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18308.cif.gz emd-18308.cif.gz | 6.3 KB | ||
| Others |  emd_18308_additional_1.map.gz emd_18308_additional_1.map.gz emd_18308_additional_2.map.gz emd_18308_additional_2.map.gz emd_18308_additional_3.map.gz emd_18308_additional_3.map.gz emd_18308_half_map_1.map.gz emd_18308_half_map_1.map.gz emd_18308_half_map_2.map.gz emd_18308_half_map_2.map.gz | 494.1 MB 495.1 MB 494.5 MB 928.8 MB 928.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18308 http://ftp.pdbj.org/pub/emdb/structures/EMD-18308 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18308 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18308 | HTTPS FTP |
-Related structure data
| Related structure data |  8qb9MC  8qb7C  8qb8C  8qbbC  8qbdC  8qbeC  8qbfC  8qbgC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18308.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18308.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Helical map Type II -PIP2/ sterol (D1, rise=5.04, twist=-136.50) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18308_msk_1.map emd_18308_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
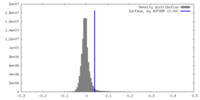
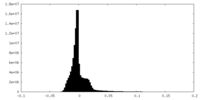
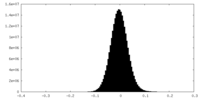
| Density Histograms |
-Additional map: Helical map Type I -PIP2/ sterol (D2, rise=9.76, twist=131.91)
| File | emd_18308_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Helical map Type I -PIP2/ sterol (D2, rise=9.76, twist=131.91) | ||||||||||||
| Projections & Slices |
| ||||||||||||
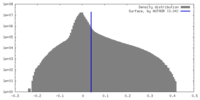
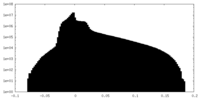
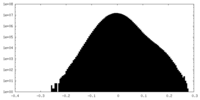
| Density Histograms |
-Additional map: Helical map Type II -PIP2/ sterol - unsharpened map
| File | emd_18308_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Helical map Type II -PIP2/ sterol - unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
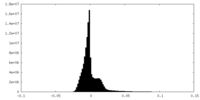
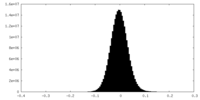
| Density Histograms |
-Additional map: Helical map Type III -PIP2/ sterol (D7, rise=36.54, twist=-14.47)
| File | emd_18308_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Helical map Type III -PIP2/ sterol (D7, rise=36.54, twist=-14.47) | ||||||||||||
| Projections & Slices |
| ||||||||||||
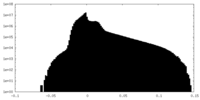
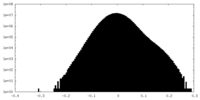
| Density Histograms |
-Half map: Helical map Type II -PIP2/ sterol - half map A
| File | emd_18308_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Helical map Type II -PIP2/ sterol - half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Helical map Type II -PIP2/ sterol - half map B
| File | emd_18308_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Helical map Type II -PIP2/ sterol - half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Helical assembly of recombinant Pil1 protein tubulating -PIP2/+st...
| Entire | Name: Helical assembly of recombinant Pil1 protein tubulating -PIP2/+sterol lipid mixture (DOPC,DOPE,DOPS,cholesterol 30:20:20:30) |
|---|---|
| Components |
|
-Supramolecule #1: Helical assembly of recombinant Pil1 protein tubulating -PIP2/+st...
| Supramolecule | Name: Helical assembly of recombinant Pil1 protein tubulating -PIP2/+sterol lipid mixture (DOPC,DOPE,DOPS,cholesterol 30:20:20:30) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Sphingolipid long chain base-responsive protein PIL1
| Macromolecule | Name: Sphingolipid long chain base-responsive protein PIL1 / type: protein_or_peptide / ID: 1 / Number of copies: 14 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.393043 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHRTYSLRNS RAPTASQLQN PPPPPSTTKG RFFGKGGLAY SFRRSAAGAF GPELSRKLSQ LVKIEKNVLR SMELTANERR DAAKQLSIW GLENDDDVSD ITDKLGVLIY EVSELDDQFI DRYDQYRLTL KSIRDIEGSV QPSRDRKDKI TDKIAYLKYK D PQSPKIEV ...String: MHRTYSLRNS RAPTASQLQN PPPPPSTTKG RFFGKGGLAY SFRRSAAGAF GPELSRKLSQ LVKIEKNVLR SMELTANERR DAAKQLSIW GLENDDDVSD ITDKLGVLIY EVSELDDQFI DRYDQYRLTL KSIRDIEGSV QPSRDRKDKI TDKIAYLKYK D PQSPKIEV LEQELVRAEA ESLVAEAQLS NITRSKLRAA FNYQFDSIIE HSEKIALIAG YGKALLELLD DSPVTPGETR PA YDGYEAS KQIIIDAESA LNEWTLDSAQ VKPTLSFKQD YEDFEPEEGE EEEEEDGQGR WSEDEQEDGQ IEEPEQEEEG AVE EHEQVG HQQSESLPQQ TTA UniProtKB: Sphingolipid long chain base-responsive protein PIL1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: 20mM HEPES, pH 7.4, 150mM KoAc, 2mM MgAc |
|---|---|
| Grid | Model: EMS Lacey Carbon / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: LACEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 291 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 5.044 Å Applied symmetry - Helical parameters - Δ&Phi: -136.5 ° Applied symmetry - Helical parameters - Axial symmetry: D1 (2x1 fold dihedral) Resolution.type: BY AUTHOR / Resolution: 3.35 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 4.1.2) / Number images used: 176005 |
|---|---|
| Startup model | Type of model: NONE |
| Final angle assignment | Type: NOT APPLICABLE / Software - Name: cryoSPARC (ver. 4.1.2) |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)