+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Structure of CUL9-RBX1 ubiquitin E3 ligase complex - hexameric assembly | |||||||||
 マップデータ マップデータ | postprocess map | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | Cullin-RING RBR E3 Ligase / LIGASE | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報cullin-RING-type E3 NEDD8 transferase / NEDD8 transferase activity / cullin-RING ubiquitin ligase complex / cellular response to chemical stress / Cul7-RING ubiquitin ligase complex / ubiquitin-dependent protein catabolic process via the C-end degron rule pathway / Loss of Function of FBXW7 in Cancer and NOTCH1 Signaling / positive regulation of protein autoubiquitination / protein neddylation / NEDD8 ligase activity ...cullin-RING-type E3 NEDD8 transferase / NEDD8 transferase activity / cullin-RING ubiquitin ligase complex / cellular response to chemical stress / Cul7-RING ubiquitin ligase complex / ubiquitin-dependent protein catabolic process via the C-end degron rule pathway / Loss of Function of FBXW7 in Cancer and NOTCH1 Signaling / positive regulation of protein autoubiquitination / protein neddylation / NEDD8 ligase activity / Cul5-RING ubiquitin ligase complex / negative regulation of response to oxidative stress / ubiquitin-ubiquitin ligase activity / SCF ubiquitin ligase complex / Cul2-RING ubiquitin ligase complex / Cul4A-RING E3 ubiquitin ligase complex / regulation of mitotic nuclear division / Cul4B-RING E3 ubiquitin ligase complex / negative regulation of type I interferon production / SCF-dependent proteasomal ubiquitin-dependent protein catabolic process / Cul3-RING ubiquitin ligase complex / Prolactin receptor signaling / protein monoubiquitination / cullin family protein binding / protein K48-linked ubiquitination / Nuclear events stimulated by ALK signaling in cancer / positive regulation of TORC1 signaling / post-translational protein modification / regulation of cellular response to insulin stimulus / Regulation of BACH1 activity / T cell activation / cellular response to amino acid stimulus / Degradation of DVL / Recognition of DNA damage by PCNA-containing replication complex / Degradation of GLI1 by the proteasome / Negative regulation of NOTCH4 signaling / GSK3B and BTRC:CUL1-mediated-degradation of NFE2L2 / Vif-mediated degradation of APOBEC3G / Hedgehog 'on' state / Degradation of GLI2 by the proteasome / GLI3 is processed to GLI3R by the proteasome / DNA Damage Recognition in GG-NER / FBXL7 down-regulates AURKA during mitotic entry and in early mitosis / RING-type E3 ubiquitin transferase / negative regulation of canonical Wnt signaling pathway / Degradation of beta-catenin by the destruction complex / Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha / Transcription-Coupled Nucleotide Excision Repair (TC-NER) / Formation of TC-NER Pre-Incision Complex / Dual Incision in GG-NER / Evasion by RSV of host interferon responses / NOTCH1 Intracellular Domain Regulates Transcription / Constitutive Signaling by NOTCH1 PEST Domain Mutants / Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants / microtubule cytoskeleton organization / Regulation of expression of SLITs and ROBOs / Formation of Incision Complex in GG-NER / Interleukin-1 signaling / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / Orc1 removal from chromatin / protein polyubiquitination / Regulation of RAS by GAPs / positive regulation of protein catabolic process / cellular response to UV / Regulation of RUNX2 expression and activity / MAPK cascade / ubiquitin protein ligase activity / KEAP1-NFE2L2 pathway / Antigen processing: Ubiquitination & Proteasome degradation / positive regulation of proteasomal ubiquitin-dependent protein catabolic process / Neddylation / ubiquitin-dependent protein catabolic process / spermatogenesis / positive regulation of canonical NF-kappaB signal transduction / proteasome-mediated ubiquitin-dependent protein catabolic process / RNA polymerase II-specific DNA-binding transcription factor binding / Potential therapeutics for SARS / molecular adaptor activity / protein ubiquitination / DNA repair / DNA damage response / ubiquitin protein ligase binding / zinc ion binding / nucleoplasm / ATP binding / nucleus / cytosol / cytoplasm 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
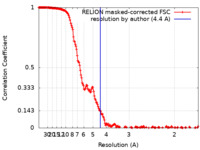
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 4.4 Å | |||||||||
 データ登録者 データ登録者 | Hopf LVM / Horn-Ghetko D / Schulman BA | |||||||||
| 資金援助 | European Union,  ドイツ, 2件 ドイツ, 2件
| |||||||||
 引用 引用 |  ジャーナル: Nat Struct Mol Biol / 年: 2024 ジャーナル: Nat Struct Mol Biol / 年: 2024タイトル: Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex. 著者: Daniel Horn-Ghetko / Linus V M Hopf / Ishita Tripathi-Giesgen / Jiale Du / Sebastian Kostrhon / D Tung Vu / Viola Beier / Barbara Steigenberger / J Rajan Prabu / Luca Stier / Elias M Bruss / ...著者: Daniel Horn-Ghetko / Linus V M Hopf / Ishita Tripathi-Giesgen / Jiale Du / Sebastian Kostrhon / D Tung Vu / Viola Beier / Barbara Steigenberger / J Rajan Prabu / Luca Stier / Elias M Bruss / Matthias Mann / Yue Xiong / Brenda A Schulman /   要旨: Ubiquitin ligation is typically executed by hallmark E3 catalytic domains. Two such domains, 'cullin-RING' and 'RBR', are individually found in several hundred human E3 ligases, and collaborate with ...Ubiquitin ligation is typically executed by hallmark E3 catalytic domains. Two such domains, 'cullin-RING' and 'RBR', are individually found in several hundred human E3 ligases, and collaborate with E2 enzymes to catalyze ubiquitylation. However, the vertebrate-specific CUL9 complex with RBX1 (also called ROC1), of interest due to its tumor suppressive interaction with TP53, uniquely encompasses both cullin-RING and RBR domains. Here, cryo-EM, biochemistry and cellular assays elucidate a 1.8-MDa hexameric human CUL9-RBX1 assembly. Within one dimeric subcomplex, an E2-bound RBR domain is activated by neddylation of its own cullin domain and positioning from the adjacent CUL9-RBX1 in trans. Our data show CUL9 as unique among RBX1-bound cullins in dependence on the metazoan-specific UBE2F neddylation enzyme, while the RBR domain protects it from deneddylation. Substrates are recruited to various upstream domains, while ubiquitylation relies on both CUL9's neddylated cullin and RBR domains achieving self-assembled and chimeric cullin-RING/RBR E3 ligase activity. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_18214.map.gz emd_18214.map.gz | 57.9 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-18214-v30.xml emd-18214-v30.xml emd-18214.xml emd-18214.xml | 21.7 KB 21.7 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_18214_fsc.xml emd_18214_fsc.xml | 17.9 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_18214.png emd_18214.png | 78.3 KB | ||
| マスクデータ |  emd_18214_msk_1.map emd_18214_msk_1.map | 488.4 MB |  マスクマップ マスクマップ | |
| Filedesc metadata |  emd-18214.cif.gz emd-18214.cif.gz | 7.2 KB | ||
| その他 |  emd_18214_additional_1.map.gz emd_18214_additional_1.map.gz emd_18214_half_map_1.map.gz emd_18214_half_map_1.map.gz emd_18214_half_map_2.map.gz emd_18214_half_map_2.map.gz | 388 MB 388 MB 388.1 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18214 http://ftp.pdbj.org/pub/emdb/structures/EMD-18214 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18214 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18214 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_18214_validation.pdf.gz emd_18214_validation.pdf.gz | 658.2 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_18214_full_validation.pdf.gz emd_18214_full_validation.pdf.gz | 657.7 KB | 表示 | |
| XML形式データ |  emd_18214_validation.xml.gz emd_18214_validation.xml.gz | 25.8 KB | 表示 | |
| CIF形式データ |  emd_18214_validation.cif.gz emd_18214_validation.cif.gz | 34.1 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18214 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18214 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18214 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18214 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8q7eMC  8q7hC  8rhzC C: 同じ文献を引用 ( M: このマップから作成された原子モデル |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_18214.map.gz / 形式: CCP4 / 大きさ: 488.4 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_18214.map.gz / 形式: CCP4 / 大きさ: 488.4 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | postprocess map | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.8512 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-マスク #1
| ファイル |  emd_18214_msk_1.map emd_18214_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-追加マップ: refinement map
| ファイル | emd_18214_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | refinement map | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: #2
| ファイル | emd_18214_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: #1
| ファイル | emd_18214_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Structure of CUL9-RBX1 ubiquitin E3 ligase complex - hexameric as...
| 全体 | 名称: Structure of CUL9-RBX1 ubiquitin E3 ligase complex - hexameric assembly |
|---|---|
| 要素 |
|
-超分子 #1: Structure of CUL9-RBX1 ubiquitin E3 ligase complex - hexameric as...
| 超分子 | 名称: Structure of CUL9-RBX1 ubiquitin E3 ligase complex - hexameric assembly タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: all |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-分子 #1: Cullin-9
| 分子 | 名称: Cullin-9 / タイプ: protein_or_peptide / ID: 1 / コピー数: 6 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 281.531875 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: MVGERHAGDL MVPLGPRLQA YPEELIRQRP GHDGHPEYLI RWSVLKCGEV GKVGVEEGKA EHILMWLSAP EVYANCPGLL GERALSKGL QHEPAGVSGS FPRDPGGLDE VAMGEMEADV QALVRRAARQ LAESGTPSLT AAVLHTIHVL SAYASIGPLT G VFRETGAL ...文字列: MVGERHAGDL MVPLGPRLQA YPEELIRQRP GHDGHPEYLI RWSVLKCGEV GKVGVEEGKA EHILMWLSAP EVYANCPGLL GERALSKGL QHEPAGVSGS FPRDPGGLDE VAMGEMEADV QALVRRAARQ LAESGTPSLT AAVLHTIHVL SAYASIGPLT G VFRETGAL DLLMHMLCNP EPQIRRSAGK MLQALAAHDA GSRAHVLLSL SQQDGIEQHM DFDSRYTLLE LFAETTSSEE HC MAFEGIH LPQIPGKLLF SLVKRYLCVT SLLDQLNSSP ELGAGDQSSP CATREKSRGQ RELEFSMAVG NLISELVRSM GWA RNLSEQ GMSPPRPTRS IFQPYISGPS LLLPTIVTTP RRQGWVFRQR SEFSSRSGYG EYVQQTLQPG MRVRMLDDYE EISA GDEGE FRQSNNGIPP VQVFWQSTGR TYWVHWHMLE ILGPEEATED KASAAVEKGA GATVLGTAFP SWDWNPMDGL YPLPY LQPE PQKNERVGYL TQAEWWELLF FIKKLDLCEQ QPIFQNLWKN LDETLGEKAL GEISVSVEMA ESLLQVLSSR FEGSTL NDL LNSQIYTKYG LLSNEPSSSS TSRNHSCTPD PEEESKSEAS FSEEETESLK AKAEAPKTEA EPTKTRTETP MAQSDSQ LF NQLLVTEGMT LPTEMKEAAS EMARALRGPG PRSSLDQHVA AVVATVQISS LDTNLQLSGL SALSQAVEEV TERDHPLV R PDRSLREKLV KMLVELLTNQ VGEKMVVVQA LRLLYLLMTK HEWRPLFARE GGIYAVLVCM QEYKTSVLVQ QAGLAALKM LAVASSSEIP TFVTGRDSIH SLFDAQMTRE IFASIDSATR PGSESLLLTV PAAVILMLNT EGCSSAARNG LLLLNLLLCN HHTLGDQII TQELRDTLFR HSGIAPRTEP MPTTRTILMM LLNRYSEPPG SPERAALETP IIQGQDGSPE LLIRSLVGGP S AELLLDLE RVLCREGSPG GAVRPLLKRL QQETQPFLLL LRTLDAPGPN KTLLLSVLRV ITRLLDFPEA MVLPWHEVLE PC LNCLSGP SSDSEIVQEL TCFLHRLASM HKDYAVVLCC LGAKEILSKV LDKHSAQLLL GCELRDLVTE CEKYAQLYSN LTS SILAGC IQMVLGQIED HRRTHQPINI PFFDVFLRHL CQGSSVEVKE DKCWEKVEVS SNPHRASKLT DHNPKTYWES NGST GSHYI TLHMHRGVLV RQLTLLVASE DSSYMPARVV VFGGDSTSCI GTELNTVNVM PSASRVILLE NLNRFWPIIQ IRIKR CQQG GIDTRVRGVE VLGPKPTFWP LFREQLCRRT CLFYTIRAQA WSRDIAEDHR RLLQLCPRLN RVLRHEQNFA DRFLPD DEA AQALGKTCWE ALVSPLVQNI TSPDAEGVSA LGWLLDQYLE QRETSRNPLS RAASFASRVR RLCHLLVHVE PPPGPSP EP STRPFSKNSK GRDRSPAPSP VLPSSSLRNI TQCWLSVVQE QVSRFLAAAW RAPDFVPRYC KLYEHLQRAG SELFGPRA A FMLALRSGFS GALLQQSFLT AAHMSEQFAR YIDQQIQGGL IGGAPGVEML GQLQRHLEPI MVLSGLELAT TFEHFYQHY MADRLLSFGS SWLEGAVLEQ IGLCFPNRLP QLMLQSLSTS EELQRQFHLF QLQRLDKLFL EQEDEEEKRL EEEEEEEEEE EAEKELFIE DPSPAISILV LSPRCWPVSP LCYLYHPRKC LPTEFCDALD RFSSFYSQSQ NHPVLDMGPH RRLQWTWLGR A ELQFGKQI LHVSTVQMWL LLKFNQTEEV SVETLLKDSD LSPELLLQAL VPLTSGNGPL TLHEGQDFPH GGVLRLHEPG PQ RSGEALW LIPPQAYLNV EKDEGRTLEQ KRNLLSCLLV RILKAHGEKG LHIDQLVCLV LEAWQKGPNP PGTLGHTVAG GVA CTSTDV LSCILHLLGQ GYVKRRDDRP QILMYAAPEP MGPCRGQADV PFCGSQSETS KPSPEAVATL ASLQLPAGRT MSPQ EVEGL MKQTVRQVQE TLNLEPDVAQ HLLAHSHWGA EQLLQSYSED PEPLLLAAGL CVHQAQAVPV RPDHCPVCVS PLGCD DDLP SLCCMHYCCK SCWNEYLTTR IEQNLVLNCT CPIADCPAQP TGAFIRAIVS SPEVISKYEK ALLRGYVESC SNLTWC TNP QGCDRILCRQ GLGCGTTCSK CGWASCFNCS FPEAHYPASC GHMSQWVDDG GYYDGMSVEA QSKHLAKLIS KRCPSCQ AP IEKNEGCLHM TCAKCNHGFC WRCLKSWKPN HKDYYNCSAM VSKAARQEKR FQDYNERCTF HHQAREFAVN LRNRVSAI H EVPPPRSFTF LNDACQGLEQ ARKVLAYACV YSFYSQDAEY MDVVEQQTEN LELHTNALQI LLEETLLRCR DLASSLRLL RADCLSTGME LLRRIQERLL AILQHSAQDF RVGLQSPSVE AWEAKGPNMP GSQPQASSGP EAEEEEEDDE DDVPEWQQDE FDEELDNDS FSYDESENLD QETFFFGDEE EDEDEAYD UniProtKB: Cullin-9 |
-分子 #2: E3 ubiquitin-protein ligase RBX1
| 分子 | 名称: E3 ubiquitin-protein ligase RBX1 / タイプ: protein_or_peptide / ID: 2 / コピー数: 6 / 光学異性体: LEVO / EC番号: RING-type E3 ubiquitin transferase |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 12.289977 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: MAAAMDVDTP SGTNSGAGKK RFEVKKWNAV ALWAWDIVVD NCAICRNHIM DLCIECQANQ ASATSEECTV AWGVCNHAFH FHCISRWLK TRQVCPLDNR EWEFQKYGH UniProtKB: E3 ubiquitin-protein ligase RBX1 |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 3 mg/mL |
|---|---|
| 緩衝液 | pH: 7.5 |
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) 平均電子線量: 60.0 e/Å2 |
| 電子線 | 加速電圧: 200 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD 最大 デフォーカス(公称値): 2.8000000000000003 µm 最小 デフォーカス(公称値): 0.7000000000000001 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)