[English] 日本語
 Yorodumi
Yorodumi- EMDB-17947: Trimeric prM/E spike of Tick-borne encephalitis virus immature pa... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Trimeric prM/E spike of Tick-borne encephalitis virus immature particle | |||||||||
 Map data Map data | Tick-borne encephalitis virus (strain Neudoerfl) immature particle E/prM spike trimer main map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | immature particle / tick-borne / flavivirus / spike trimer / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationflavivirin / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / viral capsid / double-stranded RNA binding / nucleoside-triphosphate phosphatase / clathrin-dependent endocytosis of virus by host cell / mRNA (guanine-N7)-methyltransferase / methyltransferase cap1 / methyltransferase cap1 activity ...flavivirin / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / viral capsid / double-stranded RNA binding / nucleoside-triphosphate phosphatase / clathrin-dependent endocytosis of virus by host cell / mRNA (guanine-N7)-methyltransferase / methyltransferase cap1 / methyltransferase cap1 activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / RNA helicase activity / protein dimerization activity / symbiont-mediated suppression of host innate immune response / host cell perinuclear region of cytoplasm / host cell endoplasmic reticulum membrane / RNA helicase / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / symbiont-mediated activation of host autophagy / serine-type endopeptidase activity / RNA-directed RNA polymerase / viral RNA genome replication / RNA-directed RNA polymerase activity / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell nucleus / virion membrane / structural molecule activity / ATP hydrolysis activity / proteolysis / extracellular region / ATP binding / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Tick-borne encephalitis virus Tick-borne encephalitis virus | |||||||||
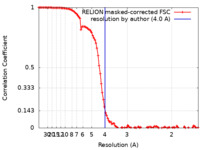
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Fuzik T / Plevka P / Smerdova L / Novacek J | |||||||||
| Funding support |  Czech Republic, 2 items Czech Republic, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: The structure of immature tick-borne encephalitis virus supports the collapse model of flavivirus maturation. Authors: Maria Anastasina / Tibor Füzik / Aušra Domanska / Lauri Ilmari Aurelius Pulkkinen / Lenka Šmerdová / Petra Pokorná Formanová / Petra Straková / Jiří Nováček / Daniel Růžek / ...Authors: Maria Anastasina / Tibor Füzik / Aušra Domanska / Lauri Ilmari Aurelius Pulkkinen / Lenka Šmerdová / Petra Pokorná Formanová / Petra Straková / Jiří Nováček / Daniel Růžek / Pavel Plevka / Sarah Jane Butcher /   Abstract: We present structures of three immature tick-borne encephalitis virus (TBEV) isolates. Our atomic models of the major viral components, the E and prM proteins, indicate that the pr domains of prM ...We present structures of three immature tick-borne encephalitis virus (TBEV) isolates. Our atomic models of the major viral components, the E and prM proteins, indicate that the pr domains of prM have a critical role in holding the heterohexameric prM3E3 spikes in a metastable conformation. Destabilization of the prM furin-sensitive loop at acidic pH facilitates its processing. The prM topology and domain assignment in TBEV is similar to the mosquito-borne Binjari virus, but is in contrast to other immature flavivirus models. These results support that prM cleavage, the collapse of E protein ectodomains onto the virion surface, the large movement of the membrane domains of both E and M, and the release of the pr fragment from the particle render the virus mature and infectious. Our work favors the collapse model of flavivirus maturation warranting further studies of immature flaviviruses to determine the sequence of events and mechanistic details driving flavivirus maturation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17947.map.gz emd_17947.map.gz | 7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17947-v30.xml emd-17947-v30.xml emd-17947.xml emd-17947.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17947_fsc.xml emd_17947_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_17947.png emd_17947.png | 121.1 KB | ||
| Masks |  emd_17947_msk_1.map emd_17947_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17947.cif.gz emd-17947.cif.gz | 6.8 KB | ||
| Others |  emd_17947_half_map_1.map.gz emd_17947_half_map_1.map.gz emd_17947_half_map_2.map.gz emd_17947_half_map_2.map.gz | 80.7 MB 80.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17947 http://ftp.pdbj.org/pub/emdb/structures/EMD-17947 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17947 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17947 | HTTPS FTP |
-Related structure data
| Related structure data |  8puvMC  8ppqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17947.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17947.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tick-borne encephalitis virus (strain Neudoerfl) immature particle E/prM spike trimer main map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8336 Å | ||||||||||||||||||||||||||||||||||||
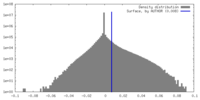
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17947_msk_1.map emd_17947_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1 of gold standard reconstruction of...
| File | emd_17947_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 of gold standard reconstruction of Tick-borne encephalitis virus (strain Neudoerfl) immature particle E/prM spike trimer | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2 of gold standard reconstruction of...
| File | emd_17947_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 of gold standard reconstruction of Tick-borne encephalitis virus (strain Neudoerfl) immature particle E/prM spike trimer | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tick-borne encephalitis virus
| Entire | Name:  Tick-borne encephalitis virus Tick-borne encephalitis virus |
|---|---|
| Components |
|
-Supramolecule #1: Tick-borne encephalitis virus
| Supramolecule | Name: Tick-borne encephalitis virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Immature particles produced using tissue cell cultures NCBI-ID: 11084 / Sci species name: Tick-borne encephalitis virus / Sci species strain: Neudoerfl / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Virus shell | Shell ID: 1 / Name: immature particle / Diameter: 600.0 Å |
-Macromolecule #1: Envelope protein E
| Macromolecule | Name: Envelope protein E / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tick-borne encephalitis virus / Strain: Neudoerfl Tick-borne encephalitis virus / Strain: Neudoerfl |
| Molecular weight | Theoretical: 53.572297 KDa |
| Sequence | String: SRCTHLENRD FVTGTQGTTR VTLVLELGGC VTITAEGKPS MDVWLDAIYQ ENPAKTREYC LHAKLSDTKV AARCPTMGPA TLAEEHQGG TVCKRDQSDR GWGNHCGLFG KGSIVACVKA ACEAKKKATG HVYDANKIVY TVKVEPHTGD YVAANETHSG R KTASFTIS ...String: SRCTHLENRD FVTGTQGTTR VTLVLELGGC VTITAEGKPS MDVWLDAIYQ ENPAKTREYC LHAKLSDTKV AARCPTMGPA TLAEEHQGG TVCKRDQSDR GWGNHCGLFG KGSIVACVKA ACEAKKKATG HVYDANKIVY TVKVEPHTGD YVAANETHSG R KTASFTIS SEKTILTMGE YGDVSLLCRV ASGVDLAQTV ILELDKTVEH LPTAWQVHRD WFNDLALPWK HEGAQNWNNA ER LVEFGAP HAVKMDVYNL GDQTGVLLKA LAGVPVAHIE GTKYHLKSGH VTCEVGLEKL KMKGLTYTMC DKTKFTWKRA PTD SGHDTV VMEVTFSGTK PCRIPVRAVA HGSPDVNVAM LITPNPTIEN NGGGFIEMQL PPGDNIIYVG ELSHQWFQKG SSIG RVFQK TKKGIERLTV IGEHAWDFGS AGGFLSSIGK AVHTVLGGAF NSIFGGVGFL PKLLLGVALA WLGLNMRNPT MSMSF LLAG GLVLAMTLGV GA UniProtKB: Genome polyprotein |
-Macromolecule #2: Protein prM
| Macromolecule | Name: Protein prM / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tick-borne encephalitis virus / Strain: Neudoerfl Tick-borne encephalitis virus / Strain: Neudoerfl |
| Molecular weight | Theoretical: 18.07667 KDa |
| Sequence | String: TVRKERDGST VIRAEGKDAA TQVRVENGTC VILATDMGSW CDDSLSYECV TIDQGEEPVD VDCFCRNVDG VYLEYGRCGK QEGSRTRRS VLIPSHAQGE LTGRGHKWLE GDSLRTHLTR VEGWVWKNKL LALAMVTVVW LTLESVVTRV AVLVVLLCLA P VYA UniProtKB: Genome polyprotein |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8.5 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 280 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | purified immature particles of Tick-borne encephalitis virus |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 11246 / Average exposure time: 2.0 sec. / Average electron dose: 40.0 e/Å2 Details: collected in movie mode total 40 fractions per micrograph |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 30.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Details | inital alhafol generated model was rigidbody fitted into map by Chimera, manually adjusted in Coot and realspace refined in Phenix |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: CC |
| Output model |  PDB-8puv: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)