[English] 日本語
 Yorodumi
Yorodumi- EMDB-17858: Tilapia Lake Virus polymerase in vRNA initiation state (core only) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tilapia Lake Virus polymerase in vRNA initiation state (core only) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Viral polymerase / VIRAL PROTEIN | |||||||||
| Function / homology | RNA-directed RNA polymerase activity / DNA/RNA polymerase superfamily / Uncharacterized protein / Putative PB1 / RNA-dependent RNA polymerase Function and homology information Function and homology information | |||||||||
| Biological species |  Tilapia lake virus / synthetic construct (others) Tilapia lake virus / synthetic construct (others) | |||||||||
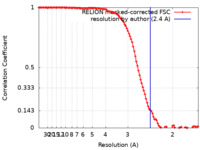
| Method | single particle reconstruction / cryo EM / Resolution: 2.4 Å | |||||||||
 Authors Authors | Arragain B / Cusack S | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural and functional analysis of the minimal orthomyxovirus-like polymerase of Tilapia Lake Virus from the highly diverged Amnoonviridae family. Authors: Benoit Arragain / Martin Pelosse / Albert Thompson / Stephen Cusack /   Abstract: Tilapia Lake Virus (TiLV), a recently discovered pathogen of tilapia fish, belongs to the Amnoonviridae family from the Articulavirales order. Its ten genome segments have characteristic conserved ...Tilapia Lake Virus (TiLV), a recently discovered pathogen of tilapia fish, belongs to the Amnoonviridae family from the Articulavirales order. Its ten genome segments have characteristic conserved ends and encode proteins with no known homologues, apart from the segment 1, which encodes an orthomyxo-like RNA-dependent-RNA polymerase core subunit. Here we show that segments 1-3 encode respectively the PB1, PB2 and PA-like subunits of an active heterotrimeric polymerase that maintains all domains found in the distantly related influenza polymerase, despite an unprecedented overall size reduction of 40%. Multiple high-resolution cryo-EM structures of TiLV polymerase in pre-initiation, initiation and active elongation states, show how it binds the vRNA and cRNA promoters and performs RNA synthesis, with both transcriptase and replicase configurations being characterised. However, the highly truncated endonuclease-like domain appears inactive and the putative cap-binding domain is autoinhibited, emphasising that many functional aspects of TiLV polymerase remain to be elucidated. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17858.map.gz emd_17858.map.gz | 7.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17858-v30.xml emd-17858-v30.xml emd-17858.xml emd-17858.xml | 21.6 KB 21.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17858_fsc.xml emd_17858_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_17858.png emd_17858.png | 57.9 KB | ||
| Masks |  emd_17858_msk_1.map emd_17858_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17858.cif.gz emd-17858.cif.gz | 6.8 KB | ||
| Others |  emd_17858_additional_1.map.gz emd_17858_additional_1.map.gz emd_17858_half_map_1.map.gz emd_17858_half_map_1.map.gz emd_17858_half_map_2.map.gz emd_17858_half_map_2.map.gz | 80.7 MB 80.7 MB 80.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17858 http://ftp.pdbj.org/pub/emdb/structures/EMD-17858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17858 | HTTPS FTP |
-Related structure data
| Related structure data |  8psoMC  8psnC  8psqC  8pssC  8psuC  8psxC  8pszC  8pt2C  8pt6C  8pt7C  8pthC  8ptjC  8qz8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17858.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17858.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17858_msk_1.map emd_17858_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
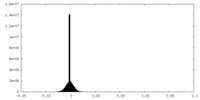
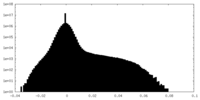
| Density Histograms |
-Additional map: #1
| File | emd_17858_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
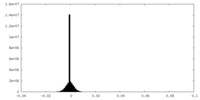
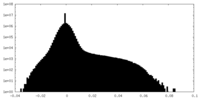
| Density Histograms |
-Half map: #1
| File | emd_17858_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17858_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tilapia lake virus polymerase
| Entire | Name: Tilapia lake virus polymerase |
|---|---|
| Components |
|
-Supramolecule #1: Tilapia lake virus polymerase
| Supramolecule | Name: Tilapia lake virus polymerase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Tilapia lake virus Tilapia lake virus |
-Macromolecule #1: Polymerase acidic protein (PA-like)
| Macromolecule | Name: Polymerase acidic protein (PA-like) / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tilapia lake virus Tilapia lake virus |
| Molecular weight | Theoretical: 47.780508 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MDSRFAQLTG VFCDDFTYSE GSRRFLSSYS TVERRPGVPV EGDCYDCLKN KWIAFELEGQ PRKFPKATVR CILNNDATYV CSEQEYQQI CKVQFKDYLE IDGVVKVGHK ASYDAELRER LLELPHPKSG PKPRIEWVAP PRLADISKET AELKRQYGFF E CSKFLACG ...String: MDSRFAQLTG VFCDDFTYSE GSRRFLSSYS TVERRPGVPV EGDCYDCLKN KWIAFELEGQ PRKFPKATVR CILNNDATYV CSEQEYQQI CKVQFKDYLE IDGVVKVGHK ASYDAELRER LLELPHPKSG PKPRIEWVAP PRLADISKET AELKRQYGFF E CSKFLACG EECGLDQEAR ELILNEYARD REFEFRNGGW IQRYTVASHK PATQKILPLP ASAPLARELL MLIARSTTQA GK VLHSDNT SILAVPVMRD SGKHSKRRPT ASTHHLVVGL SKPGCEHDFE FDGYRAAVHV MHLDPKQSAN IGEQDFVSTR EIY KLDMLE LPPISRKGDL DRASGLETRW DVILLLECLD STRVSQAVAQ HFNRHRLALS VCKDEFRKGY QLASEIRGTI PLSS LYYSL CAVRLRMTVH PFAR UniProtKB: Uncharacterized protein |
-Macromolecule #2: Putative PB1
| Macromolecule | Name: Putative PB1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tilapia lake virus Tilapia lake virus |
| Molecular weight | Theoretical: 57.179375 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MWAFQEGVCK GNLLSGPTSM KAPDSAARES IDRASEIMTG KSYNAVHTGD LSKLPNQGES PLRIVDSDLY SERSCCWVIE KEGRVVCKS TTLTRGMTSL LNTTKCSSPS ELICKVLTVE SLSEKIGDTS VEELLSHGRY FKCALRDQER GKPKSRAIFL S HPFFRLLS ...String: MWAFQEGVCK GNLLSGPTSM KAPDSAARES IDRASEIMTG KSYNAVHTGD LSKLPNQGES PLRIVDSDLY SERSCCWVIE KEGRVVCKS TTLTRGMTSL LNTTKCSSPS ELICKVLTVE SLSEKIGDTS VEELLSHGRY FKCALRDQER GKPKSRAIFL S HPFFRLLS SVVETHARSV LSKVSAVYTA TASAEQRAMM AAQVVESRKH VLNGDCTKYN EAIDADTLLK VWDAIGMGSI GV MLAYMVR RKCVLIKDTL VECPGGMLMG MFNATATLAL QGTTDRFLSF SDDFITSFNS PAELREIEDL LFASCHNLSL KKS YISVAS LEINSCTLTR DGDLATGLGC TAGVPFRGPL VTLKQTAAML SGAVDSGVMP FHSAERLFQI KQQECAYRYN NPTY TTRNE DFLPTCLGGK TVISFQSLLT WDCHPFWYQV HPDGPDTIDQ KVLSVLASKT RRRRTRLEAL SDLDPLVPHR LLVSE SDVS KIRAARQAHL KSLGLEQPTN FNYAIYKAVQ PTAGC UniProtKB: Putative PB1 |
-Macromolecule #3: RNA-dependent RNA polymerase
| Macromolecule | Name: RNA-dependent RNA polymerase / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tilapia lake virus Tilapia lake virus |
| Molecular weight | Theoretical: 53.782141 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSQFGKSFKG RTEVTITEYR SHTVKDVHRS LLTADKSLRK SFCFRNALNQ FLDKDLPLLP IRPKLESRVA VKKSKLRSQL SFRPGLTQE EAIDLYNKGY DGDSVSGALQ DRVVNEPVAY SSADNDKFHR GLAALGYTLA DRAFDTCESG FVRAIPTTPC G FICCGPGS ...String: MSQFGKSFKG RTEVTITEYR SHTVKDVHRS LLTADKSLRK SFCFRNALNQ FLDKDLPLLP IRPKLESRVA VKKSKLRSQL SFRPGLTQE EAIDLYNKGY DGDSVSGALQ DRVVNEPVAY SSADNDKFHR GLAALGYTLA DRAFDTCESG FVRAIPTTPC G FICCGPGS FKDSLGFVIK IGEFWHMYDG FQHFVAVEDA KFLASKSPSF WLAKRLAKRL NLVPKEDPSV AAAECPCKKV WE ASFARAP TALDPFGGRA FCDQGWVYHR DVGYATANHI SQETLFQQAL SVRNLGPQGS ANVSGSIHTA LDRLRAAYSR GTP ASRSIL QGLANLITPV GENFECDLDK RKLNIKALRS PERYITIEGL VVNLDDVVRG FYLDKAKVTV LSRSKWMGYE DLPQ KPPNG TFYCRKRKAM LLISCSPGTY AKKRKVAVQE DRFKDMRVEN FREVAENMDL NQGSGSENLY FQGHHHHHHH HHH UniProtKB: RNA-dependent RNA polymerase |
-Macromolecule #4: 5' vRNA end - vRNA loop (40-mer)
| Macromolecule | Name: 5' vRNA end - vRNA loop (40-mer) / type: rna / ID: 4 / Number of copies: 2 |
|---|---|
| Source (natural) | Organism:  Tilapia lake virus Tilapia lake virus |
| Molecular weight | Theoretical: 12.736518 KDa |
| Sequence | String: GCAAAUCUUU CUCACGUCCU GACUUGUGAG UAAAAUUUGG |
-Macromolecule #5: DNA (5'-D(*(CTP))-3')
| Macromolecule | Name: DNA (5'-D(*(CTP))-3') / type: dna / ID: 5 / Details: CTP / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 483.156 Da |
| Sequence | String: (CTP) |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #7: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 7 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 44 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8pso: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)