[English] 日本語
 Yorodumi
Yorodumi- EMDB-17722: Cas1-Cas2 CRISPR integrase bound to prespacer DNA, Streptococcus ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cas1-Cas2 CRISPR integrase bound to prespacer DNA, Streptococcus thermophilus DGCC 7710 CRISPR3 system | |||||||||
 Map data Map data | Map autosharpened using phenix.auto_sharpen_1.21rc1-4903 b_iso_to_d_cut and 3.17 A cutoff | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cas1-Cas2 integrase / CRISPR-Cas / prespacer / spacer acquisition / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of CRISPR repeat elements / RNA endonuclease activity / DNA endonuclease activity / defense response to virus / Hydrolases; Acting on ester bonds / DNA binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Streptococcus thermophilus DGCC 7710 (bacteria) / synthetic construct (others) Streptococcus thermophilus DGCC 7710 (bacteria) / synthetic construct (others) | |||||||||
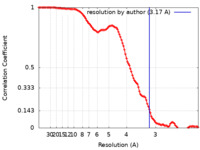
| Method | single particle reconstruction / cryo EM / Resolution: 3.17 Å | |||||||||
 Authors Authors | Sasnauskas G / Gaizauskaite U / Tamulaitiene G | |||||||||
| Funding support | Lithuania, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structural basis for spacer acquisition in a type II-A CRISPR-Cas system Authors: Sasnauskas G / Gaizauskaite U / Tamulaitiene G | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17722.map.gz emd_17722.map.gz | 112.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17722-v30.xml emd-17722-v30.xml emd-17722.xml emd-17722.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17722_fsc.xml emd_17722_fsc.xml | 14.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_17722.png emd_17722.png | 147.4 KB | ||
| Masks |  emd_17722_msk_1.map emd_17722_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17722.cif.gz emd-17722.cif.gz | 6.5 KB | ||
| Others |  emd_17722_additional_1.map.gz emd_17722_additional_1.map.gz emd_17722_half_map_1.map.gz emd_17722_half_map_1.map.gz emd_17722_half_map_2.map.gz emd_17722_half_map_2.map.gz | 62.8 MB 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17722 http://ftp.pdbj.org/pub/emdb/structures/EMD-17722 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17722 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17722 | HTTPS FTP |
-Related structure data
| Related structure data |  8pk1MC  8pj9C  8q2nC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17722.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17722.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map autosharpened using phenix.auto_sharpen_1.21rc1-4903 b_iso_to_d_cut and 3.17 A cutoff | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
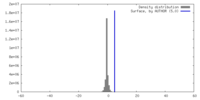
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17722_msk_1.map emd_17722_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
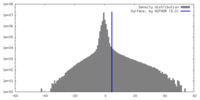
| Density Histograms |
-Additional map: unsharpened map
| File | emd_17722_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
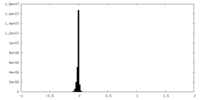
| Density Histograms |
-Half map: #2
| File | emd_17722_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
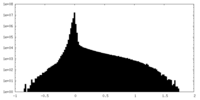
| Density Histograms |
-Half map: #1
| File | emd_17722_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cas1-Cas2 CRISPR integrase-prespacer DNA complex
| Entire | Name: Cas1-Cas2 CRISPR integrase-prespacer DNA complex |
|---|---|
| Components |
|
-Supramolecule #1: Cas1-Cas2 CRISPR integrase-prespacer DNA complex
| Supramolecule | Name: Cas1-Cas2 CRISPR integrase-prespacer DNA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Streptococcus thermophilus DGCC 7710 (bacteria) Streptococcus thermophilus DGCC 7710 (bacteria) |
-Macromolecule #1: CRISPR-associated endoribonuclease Cas2
| Macromolecule | Name: CRISPR-associated endoribonuclease Cas2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: Hydrolases; Acting on ester bonds |
|---|---|
| Source (natural) | Organism:  Streptococcus thermophilus DGCC 7710 (bacteria) Streptococcus thermophilus DGCC 7710 (bacteria) |
| Molecular weight | Theoretical: 13.43156 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSYRYMRMIL MFDMPTDTAE ERKAYRKFRK FLLSEGFIMH QFSVYSKLLL NHTANTAMVG RLKANNPKKG NITILTVTEK QFARMIYLY GDKNTSIANS EERLVFLGDN YCDED UniProtKB: CRISPR-associated endoribonuclease Cas2 |
-Macromolecule #2: CRISPR-associated endonuclease Cas1
| Macromolecule | Name: CRISPR-associated endonuclease Cas1 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO / EC number: Hydrolases; Acting on ester bonds |
|---|---|
| Source (natural) | Organism:  Streptococcus thermophilus DGCC 7710 (bacteria) Streptococcus thermophilus DGCC 7710 (bacteria) |
| Molecular weight | Theoretical: 35.363461 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAGWRTVVVN IHSKLSYKNN HLIFRNSYKT EMIHLSEIDI LLLETTDIVL TTMLVKRLVD ENILVIFCDD KRLPTAFLTP YYARHDSSL QIARQIAWKE NVKCEVWTAI IAQKILNQSY YLGECSFFEK SQSIMELYHG LERFDPSNRE GHSARIYFNT L FGNDFTRE ...String: MAGWRTVVVN IHSKLSYKNN HLIFRNSYKT EMIHLSEIDI LLLETTDIVL TTMLVKRLVD ENILVIFCDD KRLPTAFLTP YYARHDSSL QIARQIAWKE NVKCEVWTAI IAQKILNQSY YLGECSFFEK SQSIMELYHG LERFDPSNRE GHSARIYFNT L FGNDFTRE SDNDINAALD YGYTLLLSMF AREVVVCGCM TQIGLKHANQ FNQFNLASDI MEPFRPIIDR IVYQNRHNNF VK IKKELFS IFSETYLYNG KEMYLSNIVS DYTKKVIKAL NQLGEEIPEF RILESGWSHP QFEKA UniProtKB: CRISPR-associated endonuclease Cas1 |
-Macromolecule #3: Chains: G,J
| Macromolecule | Name: Chains: G,J / type: dna / ID: 3 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 7.917082 KDa |
| Sequence | String: (DT)(DT)(DT)(DA)(DC)(DT)(DA)(DC)(DT)(DC) (DG)(DT)(DT)(DC)(DT)(DG)(DG)(DT)(DG)(DT) (DT)(DT)(DC)(DT)(DC)(DG) |
-Macromolecule #4: Chains: H,K
| Macromolecule | Name: Chains: H,K / type: dna / ID: 4 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 8.062258 KDa |
| Sequence | String: (DA)(DA)(DA)(DC)(DA)(DC)(DC)(DA)(DG)(DA) (DA)(DC)(DG)(DA)(DG)(DT)(DA)(DG)(DT)(DA) (DA)(DA)(DT)(DT)(DG)(DG) |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - #0 - Film type ID: 1 / Support film - #0 - Material: CARBON / Support film - #1 - Film type ID: 2 / Support film - #1 - Material: GRAPHENE OXIDE / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 3 / Number real images: 3373 / Average exposure time: 46.33 sec. / Average electron dose: 30.5 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 92000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Output model |  PDB-8pk1: |
 Movie
Movie Controller
Controller






 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)