[English] 日本語
 Yorodumi
Yorodumi- EMDB-17528: CryoEM structure of METTL6 tRNA SerRS complex in a 1:2:2 stoichiometry -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of METTL6 tRNA SerRS complex in a 1:2:2 stoichiometry | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | METTL6 / tRNA / SerRS / Serine tRNA / 3-Methylcytosine / RNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationselenocysteine-tRNA ligase activity / tRNA (cytidine-3-)-methyltransferase activity / negative regulation of vascular endothelial growth factor production / selenocysteine incorporation / serine-tRNA ligase / serine-tRNA ligase activity / seryl-tRNA aminoacylation / Cytosolic tRNA aminoacylation / tRNA methylation / tRNA modification ...selenocysteine-tRNA ligase activity / tRNA (cytidine-3-)-methyltransferase activity / negative regulation of vascular endothelial growth factor production / selenocysteine incorporation / serine-tRNA ligase / serine-tRNA ligase activity / seryl-tRNA aminoacylation / Cytosolic tRNA aminoacylation / tRNA methylation / tRNA modification / Selenocysteine synthesis / negative regulation of angiogenesis / Transferases; Transferring one-carbon groups; Methyltransferases / cytoplasmic translation / tRNA binding / molecular adaptor activity / translation / RNA polymerase II cis-regulatory region sequence-specific DNA binding / negative regulation of transcription by RNA polymerase II / enzyme binding / protein homodimerization activity / extracellular exosome / ATP binding / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) | |||||||||
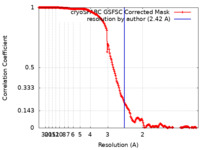
| Method | single particle reconstruction / cryo EM / Resolution: 2.42 Å | |||||||||
 Authors Authors | Throll P / Dolce LG / Kowalinski E | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structural basis of tRNA recognition by the mC RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase. Authors: Philipp Throll / Luciano G Dolce / Palma Rico-Lastres / Katharina Arnold / Laura Tengo / Shibom Basu / Stefanie Kaiser / Robert Schneider / Eva Kowalinski /   Abstract: Methylation of cytosine 32 in the anticodon loop of tRNAs to 3-methylcytosine (mC) is crucial for cellular translation fidelity. Misregulation of the RNA methyltransferases setting this modification ...Methylation of cytosine 32 in the anticodon loop of tRNAs to 3-methylcytosine (mC) is crucial for cellular translation fidelity. Misregulation of the RNA methyltransferases setting this modification can cause aggressive cancers and metabolic disturbances. Here, we report the cryo-electron microscopy structure of the human mC tRNA methyltransferase METTL6 in complex with seryl-tRNA synthetase (SerRS) and their common substrate tRNA. Through the complex structure, we identify the tRNA-binding domain of METTL6. We show that SerRS acts as the tRNA substrate selection factor for METTL6. We demonstrate that SerRS augments the methylation activity of METTL6 and that direct contacts between METTL6 and SerRS are necessary for efficient tRNA methylation. Finally, on the basis of the structure of METTL6 in complex with SerRS and tRNA, we postulate a universal tRNA-binding mode for mC RNA methyltransferases, including METTL2 and METTL8, suggesting that these mammalian paralogs use similar ways to engage their respective tRNA substrates and cofactors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17528.map.gz emd_17528.map.gz | 316.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17528-v30.xml emd-17528-v30.xml emd-17528.xml emd-17528.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17528_fsc.xml emd_17528_fsc.xml | 18.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_17528.png emd_17528.png | 33.2 KB | ||
| Masks |  emd_17528_msk_1.map emd_17528_msk_1.map | 634.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17528.cif.gz emd-17528.cif.gz | 6.8 KB | ||
| Others |  emd_17528_half_map_1.map.gz emd_17528_half_map_1.map.gz emd_17528_half_map_2.map.gz emd_17528_half_map_2.map.gz | 588.7 MB 588.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17528 http://ftp.pdbj.org/pub/emdb/structures/EMD-17528 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17528 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17528 | HTTPS FTP |
-Validation report
| Summary document |  emd_17528_validation.pdf.gz emd_17528_validation.pdf.gz | 912.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17528_full_validation.pdf.gz emd_17528_full_validation.pdf.gz | 912 KB | Display | |
| Data in XML |  emd_17528_validation.xml.gz emd_17528_validation.xml.gz | 28.2 KB | Display | |
| Data in CIF |  emd_17528_validation.cif.gz emd_17528_validation.cif.gz | 37.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17528 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17528 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17528 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17528 | HTTPS FTP |
-Related structure data
| Related structure data |  8p7bMC  8owxC  8owyC  8p7cC  8p7dC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17528.map.gz / Format: CCP4 / Size: 634.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17528.map.gz / Format: CCP4 / Size: 634.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.645 Å | ||||||||||||||||||||||||||||||||||||
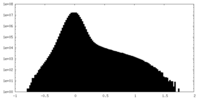
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17528_msk_1.map emd_17528_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
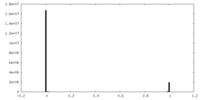
| Density Histograms |
-Half map: #2
| File | emd_17528_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
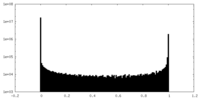
| Density Histograms |
-Half map: #1
| File | emd_17528_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
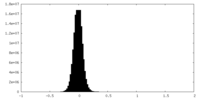
| Density Histograms |
- Sample components
Sample components
-Entire : METTL6 tRNA SerRS complex in a 1:2:2 stoichiometry
| Entire | Name: METTL6 tRNA SerRS complex in a 1:2:2 stoichiometry |
|---|---|
| Components |
|
-Supramolecule #1: METTL6 tRNA SerRS complex in a 1:2:2 stoichiometry
| Supramolecule | Name: METTL6 tRNA SerRS complex in a 1:2:2 stoichiometry / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Serine--tRNA ligase, cytoplasmic
| Macromolecule | Name: Serine--tRNA ligase, cytoplasmic / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: serine-tRNA ligase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 58.863211 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MVLDLDLFRV DKGGDPALIR ETQEKRFKDP GLVDQLVKAD SEWRRCRFRA DNLNKLKNLC SKTIGEKMKK KEPVGDDESV PENVLSFDD LTADALANLK VSQIKKVRLL IDEAILKCDA ERIKLEAERF ENLREIGNLL HPSVPISNDE DVDNKVERIW G DCTVRKKY ...String: MVLDLDLFRV DKGGDPALIR ETQEKRFKDP GLVDQLVKAD SEWRRCRFRA DNLNKLKNLC SKTIGEKMKK KEPVGDDESV PENVLSFDD LTADALANLK VSQIKKVRLL IDEAILKCDA ERIKLEAERF ENLREIGNLL HPSVPISNDE DVDNKVERIW G DCTVRKKY SHVDLVVMVD GFEGEKGAVV AGSRGYFLKG VLVFLEQALI QYALRTLGSR GYIPIYTPFF MRKEVMQEVA QL SQFDEEL YKVIGKGSEK SDDNSYDEKY LIATSEQPIA ALHRDEWLRP EDLPIKYAGL STCFRQEVGS HGRDTRGIFR VHQ FEKIEQ FVYSSPHDNK SWEMFEEMIT TAEEFYQSLG IPYHIVNIVS GSLNHAASKK LDLEAWFPGS GAFRELVSCS NCTD YQARR LRIRYGQTKK MMDKVEFVHM LNATMCATTR TICAILENYQ TEKGITVPEK LKEFMPPGLQ ELIPFVKPAP IEQEP SKKQ KKQHEGSKKK AAARDVTLEN RLQNMEVTDA UniProtKB: Serine--tRNA ligase, cytoplasmic |
-Macromolecule #2: tRNA N(3)-methylcytidine methyltransferase METTL6
| Macromolecule | Name: tRNA N(3)-methylcytidine methyltransferase METTL6 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number: Transferases; Transferring one-carbon groups; Methyltransferases |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 33.296055 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASLQRKGLQ ARILTSEEEE KLKRDQTLVS DFKQQKLEQE AQKNWDLFYK RNSTNFFKDR HWTTREFEEL RSCREFEDQK LTMLEAGCG VGNCLFPLLE EDPNIFAYAC DFSPRAIEYV KQNPLYDTER CKVFQCDLTK DDLLDHVPPE SVDVVMLIFV L SAVHPDKM ...String: MASLQRKGLQ ARILTSEEEE KLKRDQTLVS DFKQQKLEQE AQKNWDLFYK RNSTNFFKDR HWTTREFEEL RSCREFEDQK LTMLEAGCG VGNCLFPLLE EDPNIFAYAC DFSPRAIEYV KQNPLYDTER CKVFQCDLTK DDLLDHVPPE SVDVVMLIFV L SAVHPDKM HLVLQNIYKV LKPGKSVLFR DYGLYDHAML RFKASSKLGE NFYVRQDGTR SYFFTDDFLA QLFMDTGYEE VV NEYVFRE TVNKKEGLCV PRVFLQSKFL KPPKNPSPVV LGLDPKS UniProtKB: tRNA N(3)-methylcytidine methyltransferase METTL6 |
-Macromolecule #3: Serine tRNA
| Macromolecule | Name: Serine tRNA / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Molecular weight | Theoretical: 27.536477 KDa |
| Sequence | String: GCAGUGGUGG C(4AC)GAGU(OMG)G(H2U)U AAGGC(M2G)UCGG ACUUGAAA(PSU)C CGA(OMU)UCGCUC UGCGAG (5MC)GU GGG(5MU)(PSU)CG(1MA)AU CCCACCCACU GCGCCA |
-Macromolecule #4: Serine tRNA
| Macromolecule | Name: Serine tRNA / type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Molecular weight | Theoretical: 27.620635 KDa |
| Sequence | String: GCAGUGGUGG C(4AC)GAGU(OMG)G(H2U)U AAGGC(M2G)UCGG A(JMH)UUGA(6IA)A(PSU)C CGA(OMU)UCGC U CUGCGAG(5MC)GU GGG(5MU)(PSU)CG(1MA)AU CCCACCCACU GCGCCA |
-Macromolecule #5: S-ADENOSYL-L-HOMOCYSTEINE
| Macromolecule | Name: S-ADENOSYL-L-HOMOCYSTEINE / type: ligand / ID: 5 / Number of copies: 1 / Formula: SAH |
|---|---|
| Molecular weight | Theoretical: 384.411 Da |
| Chemical component information |  ChemComp-SAH: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 2 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 63.27 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)