+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Serine tRNA from Trichoplusia ni | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Serine tRNA / RNA BINDING PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) | |||||||||
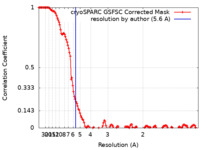
| Method | single particle reconstruction / cryo EM / Resolution: 5.6 Å | |||||||||
 Authors Authors | Throll P / Dolce LG / Kowalinski E | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structural basis of tRNA recognition by the mC RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase. Authors: Philipp Throll / Luciano G Dolce / Palma Rico-Lastres / Katharina Arnold / Laura Tengo / Shibom Basu / Stefanie Kaiser / Robert Schneider / Eva Kowalinski /   Abstract: Methylation of cytosine 32 in the anticodon loop of tRNAs to 3-methylcytosine (mC) is crucial for cellular translation fidelity. Misregulation of the RNA methyltransferases setting this modification ...Methylation of cytosine 32 in the anticodon loop of tRNAs to 3-methylcytosine (mC) is crucial for cellular translation fidelity. Misregulation of the RNA methyltransferases setting this modification can cause aggressive cancers and metabolic disturbances. Here, we report the cryo-electron microscopy structure of the human mC tRNA methyltransferase METTL6 in complex with seryl-tRNA synthetase (SerRS) and their common substrate tRNA. Through the complex structure, we identify the tRNA-binding domain of METTL6. We show that SerRS acts as the tRNA substrate selection factor for METTL6. We demonstrate that SerRS augments the methylation activity of METTL6 and that direct contacts between METTL6 and SerRS are necessary for efficient tRNA methylation. Finally, on the basis of the structure of METTL6 in complex with SerRS and tRNA, we postulate a universal tRNA-binding mode for mC RNA methyltransferases, including METTL2 and METTL8, suggesting that these mammalian paralogs use similar ways to engage their respective tRNA substrates and cofactors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17532.map.gz emd_17532.map.gz | 117 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17532-v30.xml emd-17532-v30.xml emd-17532.xml emd-17532.xml | 12.5 KB 12.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17532_fsc.xml emd_17532_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_17532.png emd_17532.png | 16.2 KB | ||
| Masks |  emd_17532_msk_1.map emd_17532_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17532.cif.gz emd-17532.cif.gz | 4.3 KB | ||
| Others |  emd_17532_half_map_1.map.gz emd_17532_half_map_1.map.gz emd_17532_half_map_2.map.gz emd_17532_half_map_2.map.gz | 226.6 MB 226.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17532 http://ftp.pdbj.org/pub/emdb/structures/EMD-17532 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17532 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17532 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17532.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17532.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.645 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17532_msk_1.map emd_17532_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17532_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17532_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : METTL6 tRNA SerRS complex in a 1:2:2 stoichiometry
| Entire | Name: METTL6 tRNA SerRS complex in a 1:2:2 stoichiometry |
|---|---|
| Components |
|
-Supramolecule #1: METTL6 tRNA SerRS complex in a 1:2:2 stoichiometry
| Supramolecule | Name: METTL6 tRNA SerRS complex in a 1:2:2 stoichiometry / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Serine tRNA
| Macromolecule | Name: Serine tRNA / type: rna / ID: 1 |
|---|---|
| Source (natural) | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GCAGUGGUGG C(4AC)GAGU(OMG)G(H2U)U AAGGC(M2G)UCGG A(JMH)UUGA(6IA)A(PSU)C CGA(OMU)UCGCUC UGCGAG(5MC)GUG GG(5MU)(PSU)CG(1MA)AUC CCACCCACUG CGCCA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 63.27 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)