[English] 日本語
 Yorodumi
Yorodumi- EMDB-17172: Ternary structure of intramolecular bivalent glue degrader IBG1 b... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Ternary structure of intramolecular bivalent glue degrader IBG1 bound to BRD4 and DCAF16:DDB1deltaBPB | ||||||||||||||||||||||||||||||
 Map data Map data | Sharpened map | ||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||
 Keywords Keywords | Bromodomain / BET protein / DCAF16 / Bivalent glue / Targeted protein degradation / TPD / TRANSCRIPTION | ||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation by virus of viral protein levels in host cell / spindle assembly involved in female meiosis / epigenetic programming in the zygotic pronuclei / UV-damage excision repair / biological process involved in interaction with symbiont / regulation of mitotic cell cycle phase transition / WD40-repeat domain binding / Cul4A-RING E3 ubiquitin ligase complex / Cul4-RING E3 ubiquitin ligase complex / Cul4B-RING E3 ubiquitin ligase complex ...positive regulation by virus of viral protein levels in host cell / spindle assembly involved in female meiosis / epigenetic programming in the zygotic pronuclei / UV-damage excision repair / biological process involved in interaction with symbiont / regulation of mitotic cell cycle phase transition / WD40-repeat domain binding / Cul4A-RING E3 ubiquitin ligase complex / Cul4-RING E3 ubiquitin ligase complex / Cul4B-RING E3 ubiquitin ligase complex / ubiquitin ligase complex scaffold activity / RNA polymerase II C-terminal domain binding / negative regulation of reproductive process / negative regulation of developmental process / P-TEFb complex binding / negative regulation of DNA damage checkpoint / histone H4 reader activity / viral release from host cell / cullin family protein binding / host-mediated suppression of viral transcription / positive regulation of G2/M transition of mitotic cell cycle / positive regulation of T-helper 17 cell lineage commitment / ectopic germ cell programmed cell death / positive regulation of viral genome replication / proteasomal protein catabolic process / : / RNA polymerase II CTD heptapeptide repeat kinase activity / positive regulation of gluconeogenesis / condensed nuclear chromosome / transcription coregulator activity / nucleotide-excision repair / positive regulation of transcription elongation by RNA polymerase II / Recognition of DNA damage by PCNA-containing replication complex / regulation of circadian rhythm / DNA Damage Recognition in GG-NER / Dual Incision in GG-NER / Transcription-Coupled Nucleotide Excision Repair (TC-NER) / Formation of TC-NER Pre-Incision Complex / Formation of Incision Complex in GG-NER / Wnt signaling pathway / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / p53 binding / positive regulation of protein catabolic process / cellular response to UV / rhythmic process / site of double-strand break / chromosome / Neddylation / regulation of inflammatory response / ubiquitin-dependent protein catabolic process / histone binding / protein-macromolecule adaptor activity / Potential therapeutics for SARS / proteasome-mediated ubiquitin-dependent protein catabolic process / damaged DNA binding / transcription coactivator activity / chromosome, telomeric region / positive regulation of canonical NF-kappaB signal transduction / transcription cis-regulatory region binding / protein ubiquitination / chromatin remodeling / DNA repair / protein serine/threonine kinase activity / apoptotic process / DNA damage response / chromatin binding / regulation of transcription by RNA polymerase II / negative regulation of apoptotic process / chromatin / positive regulation of DNA-templated transcription / protein-containing complex binding / nucleolus / enzyme binding / positive regulation of transcription by RNA polymerase II / protein-containing complex / extracellular space / DNA binding / extracellular exosome / nucleoplasm / nucleus / cytoplasm Similarity search - Function | ||||||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||||||||
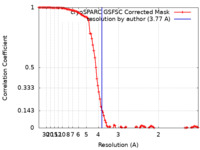
| Method | single particle reconstruction / cryo EM / Resolution: 3.77 Å | ||||||||||||||||||||||||||||||
 Authors Authors | Cowan AD / Sundaramoorthy R / Nakasone MA / Ciulli A | ||||||||||||||||||||||||||||||
| Funding support | European Union,  Switzerland, Switzerland,  United Kingdom, United Kingdom,  Austria, 9 items Austria, 9 items
| ||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Targeted protein degradation via intramolecular bivalent glues. Authors: Oliver Hsia / Matthias Hinterndorfer / Angus D Cowan / Kentaro Iso / Tasuku Ishida / Ramasubramanian Sundaramoorthy / Mark A Nakasone / Hana Imrichova / Caroline Schätz / Andrea Rukavina / ...Authors: Oliver Hsia / Matthias Hinterndorfer / Angus D Cowan / Kentaro Iso / Tasuku Ishida / Ramasubramanian Sundaramoorthy / Mark A Nakasone / Hana Imrichova / Caroline Schätz / Andrea Rukavina / Koraljka Husnjak / Martin Wegner / Alejandro Correa-Sáez / Conner Craigon / Ryan Casement / Chiara Maniaci / Andrea Testa / Manuel Kaulich / Ivan Dikic / Georg E Winter / Alessio Ciulli /     Abstract: Targeted protein degradation is a pharmacological modality that is based on the induced proximity of an E3 ubiquitin ligase and a target protein to promote target ubiquitination and proteasomal ...Targeted protein degradation is a pharmacological modality that is based on the induced proximity of an E3 ubiquitin ligase and a target protein to promote target ubiquitination and proteasomal degradation. This has been achieved either via proteolysis-targeting chimeras (PROTACs)-bifunctional compounds composed of two separate moieties that individually bind the target and E3 ligase, or via molecular glues that monovalently bind either the ligase or the target. Here, using orthogonal genetic screening, biophysical characterization and structural reconstitution, we investigate the mechanism of action of bifunctional degraders of BRD2 and BRD4, termed intramolecular bivalent glues (IBGs), and find that instead of connecting target and ligase in trans as PROTACs do, they simultaneously engage and connect two adjacent domains of the target protein in cis. This conformational change 'glues' BRD4 to the E3 ligases DCAF11 or DCAF16, leveraging intrinsic target-ligase affinities that do not translate to BRD4 degradation in the absence of compound. Structural insights into the ternary BRD4-IBG1-DCAF16 complex guided the rational design of improved degraders of low picomolar potency. We thus introduce a new modality in targeted protein degradation, which works by bridging protein domains in cis to enhance surface complementarity with E3 ligases for productive ubiquitination and degradation. #1:  Journal: Biorxiv / Year: 2023 Journal: Biorxiv / Year: 2023Title: Targeted protein degradation via intramolecular bivalent glues Authors: Hsia O / Hinterndorfer M / Cowan AD / Iso K / Ishida I / Sundaramoorthy R / Nakasone MA / Imrichova H / Schatz C / Rukvina A / Husnjak K / Wegner M / Correa-Saez A / Craigon C / Casement R / ...Authors: Hsia O / Hinterndorfer M / Cowan AD / Iso K / Ishida I / Sundaramoorthy R / Nakasone MA / Imrichova H / Schatz C / Rukvina A / Husnjak K / Wegner M / Correa-Saez A / Craigon C / Casement R / Maniaci C / Testa A / Kaulich M / Dikic I / Winter GE / Ciulli A | ||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17172.map.gz emd_17172.map.gz | 122.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17172-v30.xml emd-17172-v30.xml emd-17172.xml emd-17172.xml | 29.2 KB 29.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17172_fsc.xml emd_17172_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_17172.png emd_17172.png | 55.8 KB | ||
| Masks |  emd_17172_msk_1.map emd_17172_msk_1.map | 129.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17172.cif.gz emd-17172.cif.gz | 7.4 KB | ||
| Others |  emd_17172_additional_1.map.gz emd_17172_additional_1.map.gz emd_17172_half_map_1.map.gz emd_17172_half_map_1.map.gz emd_17172_half_map_2.map.gz emd_17172_half_map_2.map.gz | 64.6 MB 120.3 MB 120.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17172 http://ftp.pdbj.org/pub/emdb/structures/EMD-17172 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17172 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17172 | HTTPS FTP |
-Related structure data
| Related structure data |  8ov6MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17172.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17172.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.74 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17172_msk_1.map emd_17172_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
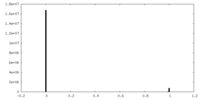
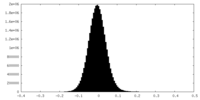
| Density Histograms |
-Additional map: Unsharpened map
| File | emd_17172_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
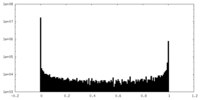
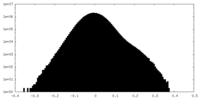
| Density Histograms |
-Half map: Half map A
| File | emd_17172_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
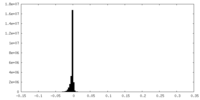
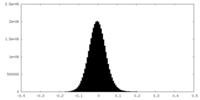
| Density Histograms |
-Half map: Half map B
| File | emd_17172_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
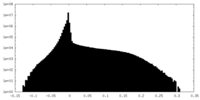
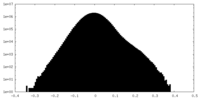
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of BRD4 tandem bromodomains, compound 1, DCAF16 a...
| Entire | Name: Ternary complex of BRD4 tandem bromodomains, compound 1, DCAF16 and DDB1deltaBPB |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of BRD4 tandem bromodomains, compound 1, DCAF16 a...
| Supramolecule | Name: Ternary complex of BRD4 tandem bromodomains, compound 1, DCAF16 and DDB1deltaBPB type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 118 KDa |
-Supramolecule #2: DCAF16:DDB1deltaBPB E3 receptor:adaptor complex
| Supramolecule | Name: DCAF16:DDB1deltaBPB E3 receptor:adaptor complex / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: DDB1deltaBPB
| Macromolecule | Name: DDB1deltaBPB / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSYNYVVTAQ KPTAVNGCVT GHFTSAEDLN LLIAKNTRLE IYVVTAEGLR PVKEVGMYGK IAVMELFRPK GESKDLLFIL TAKYNACILE YKQSGESIDI ITRAHGNVQD RIGRPSETGI IGIIDPECRM IGLRLYDGLF KVIPLDRDNK ELKAFNIRLE ELHVIDVKFL ...String: MSYNYVVTAQ KPTAVNGCVT GHFTSAEDLN LLIAKNTRLE IYVVTAEGLR PVKEVGMYGK IAVMELFRPK GESKDLLFIL TAKYNACILE YKQSGESIDI ITRAHGNVQD RIGRPSETGI IGIIDPECRM IGLRLYDGLF KVIPLDRDNK ELKAFNIRLE ELHVIDVKFL YGCQAPTICF VYQDPQGRHV KTYEVSLREK EFNKGPWKQE NVEAEASMVI AVPEPFGGAI IIGQESITYH NGDKYLAIAP PIIKQSTIVC HNRVDPNGSR YLLGDMEGRL FMLLLEKEEQ MDGTVTLKDL RVELLGETSI AECLTYLDNG VVFVGSRLGD SQLVKLNVDS NEQGSYVVAM ETFTNLGPIV DMCVVDLERQ GQGQLVTCSG AFKEGSLRII RNGIGGNGNS GEIQKLHIRT VPLYESPRKI CYQEVSQCFG VLSSRIEVQD TSGGTTALRP SASTQALSSS VSSSKLFSSS TAPHETSFGE EVEVHNLLII DQHTFEVLHA HQFLQNEYAL SLVSCKLGKD PNTYFIVGTA MVYPEEAEPK QGRIVVFQYS DGKLQTVAEK EVKGAVYSMV EFNGKLLASI NSTVRLYEWT TEKELRTECN HYNNIMALYL KTKGDFILVG DLMRSVLLLA YKPMEGNFEE IARDFNPNWM SAVEILDDDN FLGAENAFNL FVCQKDSAAT TDEERQHLQE VGLFHLGEFV NVFCHGSLVM QNLGETSTPT QGSVLFGTVN GMIGLVTSLS ESWYNLLLDM QNRLNKVIKS VGKIEHSFWR SFHTERKTEP ATGFIDGDLI ESFLDISRPK MQEVVANLQY DDGSGMKREA TADDLIKVVE ELTRIH UniProtKB: DNA damage-binding protein 1 |
-Macromolecule #2: DCAF16
| Macromolecule | Name: DCAF16 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GGMGPRNPSP DHLSESESEE EENISYLNES SGEEWDSSEE EDSMVPNLSP LESLAWQVKC LLKYSTTWKP LNPNSWLYHA KLLDPSTPVH ILREIGLRLS HCSHCVPKLE PIPEWPPLAS CGVPPFQKPL TSPSRLSRDH ATLNGALQFA TKQLSRTLSR ATPIPEYLKQ ...String: GGMGPRNPSP DHLSESESEE EENISYLNES SGEEWDSSEE EDSMVPNLSP LESLAWQVKC LLKYSTTWKP LNPNSWLYHA KLLDPSTPVH ILREIGLRLS HCSHCVPKLE PIPEWPPLAS CGVPPFQKPL TSPSRLSRDH ATLNGALQFA TKQLSRTLSR ATPIPEYLKQ IPNSCVSGCC CGWLTKTVKE TTRTEPINTT YSYTDFQKAV NKLLTASL UniProtKB: DDB1- and CUL4-associated factor 16 |
-Macromolecule #3: BRD4 Tandem Bromodomains
| Macromolecule | Name: BRD4 Tandem Bromodomains / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GSTNPPPPET SNPNKPKRQT NQLQYLLRVV LKTLWKHQFA WPFQQPVDAV KLNLPDYYKI IKTPMDMGTI KKRLENNYYW NAQECIQDFN TMFTNCYIYN KPGDDIVLMA EALEKLFLQK INELPTEETE IMIVQAKGRG RGRKETGTAK PGVSTVPNTT QASTPPQTQT ...String: GSTNPPPPET SNPNKPKRQT NQLQYLLRVV LKTLWKHQFA WPFQQPVDAV KLNLPDYYKI IKTPMDMGTI KKRLENNYYW NAQECIQDFN TMFTNCYIYN KPGDDIVLMA EALEKLFLQK INELPTEETE IMIVQAKGRG RGRKETGTAK PGVSTVPNTT QASTPPQTQT PQPNPPPVQA TPHPFPAVTP DLIVQTPVMT VVPPQPLQTP PPVPPQPQPP PAPAPQPVQS HPPIIAATPQ PVKTKKGVKR KADTTTPTTI DPIHEPPSLP PEPKTTKLGQ RRESSRPVKP PKKDVPDSQQ HPAPEKSSKV SEQLKCCSGI LKEMFAKKHA AYAWPFYKPV DVEALGLHDY CDIIKHPMDM STIKSKLEAR EYRDAQEFGA DVRLMFSNCY KYNPPDHEVV AMARKLQDVF EMRFAKMPD UniProtKB: Bromodomain-containing protein 4 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Details: Current 35 mA | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: 3.5 uL complex was dispensed onto the grid, allowed to disperse for 10 s, blotted for 3.5 s using blot force 3, then plunged into liquid ethane. | ||||||||||||
| Details | The components were co-incubated then loaded onto a 10/300 Superdex 200 gl column and the fraction corresponding to the complex was collected and concentrated to 0.8 mg/mL |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Number grids imaged: 1 / Number real images: 2075 / Average exposure time: 2.0 sec. / Average electron dose: 12.7 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 190000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.2 µm / Nominal defocus min: 1.7 µm / Nominal magnification: 190000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Output model |  PDB-8ov6: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)