[English] 日本語
 Yorodumi
Yorodumi- PDB-8ov6: Ternary structure of intramolecular bivalent glue degrader IBG1 b... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8ov6 | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
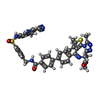
| Title | Ternary structure of intramolecular bivalent glue degrader IBG1 bound to BRD4 and DCAF16:DDB1deltaBPB | ||||||||||||||||||||||||||||||
 Components Components |
| ||||||||||||||||||||||||||||||
 Keywords Keywords | TRANSCRIPTION / Bromodomain / BET protein / DCAF16 / Bivalent glue / Targeted protein degradation / TPD | ||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationCul4-RING E3 ubiquitin ligase complex / RNA polymerase II C-terminal domain binding / P-TEFb complex binding / negative regulation of DNA damage checkpoint / histone H4 reader activity / host-mediated suppression of viral transcription / positive regulation of G2/M transition of mitotic cell cycle / positive regulation of T-helper 17 cell lineage commitment / : / RNA polymerase II CTD heptapeptide repeat kinase activity ...Cul4-RING E3 ubiquitin ligase complex / RNA polymerase II C-terminal domain binding / P-TEFb complex binding / negative regulation of DNA damage checkpoint / histone H4 reader activity / host-mediated suppression of viral transcription / positive regulation of G2/M transition of mitotic cell cycle / positive regulation of T-helper 17 cell lineage commitment / : / RNA polymerase II CTD heptapeptide repeat kinase activity / condensed nuclear chromosome / transcription coregulator activity / positive regulation of transcription elongation by RNA polymerase II / p53 binding / chromosome / Neddylation / regulation of inflammatory response / histone binding / Potential therapeutics for SARS / transcription coactivator activity / positive regulation of canonical NF-kappaB signal transduction / transcription cis-regulatory region binding / protein ubiquitination / chromatin remodeling / protein serine/threonine kinase activity / DNA damage response / chromatin binding / regulation of transcription by RNA polymerase II / chromatin / positive regulation of DNA-templated transcription / enzyme binding / positive regulation of transcription by RNA polymerase II / nucleoplasm / nucleus Similarity search - Function | ||||||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.77 Å | ||||||||||||||||||||||||||||||
 Authors Authors | Cowan, A.D. / Sundaramoorthy, R. / Nakasone, M.A. / Ciulli, A. | ||||||||||||||||||||||||||||||
| Funding support | European Union,  Switzerland, Switzerland,  United Kingdom, United Kingdom,  Austria, 9items Austria, 9items
| ||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Targeted protein degradation via intramolecular bivalent glues. Authors: Oliver Hsia / Matthias Hinterndorfer / Angus D Cowan / Kentaro Iso / Tasuku Ishida / Ramasubramanian Sundaramoorthy / Mark A Nakasone / Hana Imrichova / Caroline Schätz / Andrea Rukavina / ...Authors: Oliver Hsia / Matthias Hinterndorfer / Angus D Cowan / Kentaro Iso / Tasuku Ishida / Ramasubramanian Sundaramoorthy / Mark A Nakasone / Hana Imrichova / Caroline Schätz / Andrea Rukavina / Koraljka Husnjak / Martin Wegner / Alejandro Correa-Sáez / Conner Craigon / Ryan Casement / Chiara Maniaci / Andrea Testa / Manuel Kaulich / Ivan Dikic / Georg E Winter / Alessio Ciulli /     Abstract: Targeted protein degradation is a pharmacological modality that is based on the induced proximity of an E3 ubiquitin ligase and a target protein to promote target ubiquitination and proteasomal ...Targeted protein degradation is a pharmacological modality that is based on the induced proximity of an E3 ubiquitin ligase and a target protein to promote target ubiquitination and proteasomal degradation. This has been achieved either via proteolysis-targeting chimeras (PROTACs)-bifunctional compounds composed of two separate moieties that individually bind the target and E3 ligase, or via molecular glues that monovalently bind either the ligase or the target. Here, using orthogonal genetic screening, biophysical characterization and structural reconstitution, we investigate the mechanism of action of bifunctional degraders of BRD2 and BRD4, termed intramolecular bivalent glues (IBGs), and find that instead of connecting target and ligase in trans as PROTACs do, they simultaneously engage and connect two adjacent domains of the target protein in cis. This conformational change 'glues' BRD4 to the E3 ligases DCAF11 or DCAF16, leveraging intrinsic target-ligase affinities that do not translate to BRD4 degradation in the absence of compound. Structural insights into the ternary BRD4-IBG1-DCAF16 complex guided the rational design of improved degraders of low picomolar potency. We thus introduce a new modality in targeted protein degradation, which works by bridging protein domains in cis to enhance surface complementarity with E3 ligases for productive ubiquitination and degradation. #1:  Journal: Biorxiv / Year: 2023 Journal: Biorxiv / Year: 2023Title: Targeted protein degradation via intramolecular bivalent glues Authors: Hsia, O. / Hinterndorfer, M. / Cowan, A.D. / Iso, K. / Ishida, I. / Sundaramoorthy, R. / Nakasone, M.A. / Imrichova, H. / Schatz, C. / Rukvina, A. / Husnjak, K. / Wegner, M. / Correa-Saez, A. ...Authors: Hsia, O. / Hinterndorfer, M. / Cowan, A.D. / Iso, K. / Ishida, I. / Sundaramoorthy, R. / Nakasone, M.A. / Imrichova, H. / Schatz, C. / Rukvina, A. / Husnjak, K. / Wegner, M. / Correa-Saez, A. / Craigon, C. / Casement, R. / Maniaci, C. / Testa, A. / Kaulich, M. / Dikic, I. / Winter, G.E. / Ciulli, A. | ||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8ov6.cif.gz 8ov6.cif.gz | 482.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8ov6.ent.gz pdb8ov6.ent.gz | 324.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8ov6.json.gz 8ov6.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ov/8ov6 https://data.pdbj.org/pub/pdb/validation_reports/ov/8ov6 ftp://data.pdbj.org/pub/pdb/validation_reports/ov/8ov6 ftp://data.pdbj.org/pub/pdb/validation_reports/ov/8ov6 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  17172MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 93347.078 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: DDB1 / Production host: Homo sapiens (human) / Gene: DDB1 / Production host:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
|---|---|
| #2: Protein | Mass: 24333.449 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: DCAF16, C4orf30 / Production host: Homo sapiens (human) / Gene: DCAF16, C4orf30 / Production host:  Trichoplusia ni (cabbage looper) / References: UniProt: Q9NXF7 Trichoplusia ni (cabbage looper) / References: UniProt: Q9NXF7 |
| #3: Protein | Mass: 47203.234 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: BRD4, HUNK1 / Production host: Homo sapiens (human) / Gene: BRD4, HUNK1 / Production host:  |
| #4: Chemical | ChemComp-ZN / |
| #5: Chemical | ChemComp-U79 / |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component |
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight |
| |||||||||||||||||||||
| Source (natural) |
| |||||||||||||||||||||
| Source (recombinant) |
| |||||||||||||||||||||
| Buffer solution | pH: 7.5 | |||||||||||||||||||||
| Buffer component |
| |||||||||||||||||||||
| Specimen | Conc.: 0.8 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES Details: The components were co-incubated then loaded onto a 10/300 Superdex 200 gl column and the fraction corresponding to the complex was collected and concentrated to 0.8 mg/mL | |||||||||||||||||||||
| Specimen support | Details: Current 35 mA / Grid material: GOLD / Grid mesh size: 400 divisions/in. / Grid type: Quantifoil R1.2/1.3 | |||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K Details: 3.5 uL complex was dispensed onto the grid, allowed to disperse for 10 s, blotted for 3.5 s using blot force 3, then plunged into liquid ethane |
- Electron microscopy imaging
Electron microscopy imaging
| Microscopy | Model: TFS GLACIOS |
|---|---|
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 190000 X / Calibrated magnification: 190000 X / Nominal defocus max: 3200 nm / Nominal defocus min: 1700 nm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 2 sec. / Electron dose: 12.7 e/Å2 / Film or detector model: TFS FALCON 4i (4k x 4x) |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.77 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 192014 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | 3D fitting-ID: 1
| ||||||||||||||||||||||||||||||||||||||||||||
| Refinement | Cross valid method: NONE Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2 | ||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 100.04 Å2 | ||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj