+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Molecular Mechanism of trypanosomal AQP2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Aquaporin / Tetramer / Drug Uptake / Glycerol / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationglycerol channel activity / urea transmembrane transport / urea transmembrane transporter activity / glycerol transmembrane transport / water transport / water channel activity / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
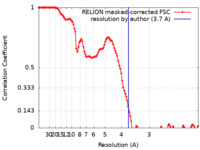
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Weyand SN / Matusevicius M / Yamashita K | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2025 Journal: Elife / Year: 2025Title: Insights from aquaporin structures into drug-resistant sleeping sickness Authors: Matusevicius M / Corey RA / Gragera M / Yamashita K / Sprenger T / Ungogo MA / Blaza JN / Castro-Hartmann P / Chirgadze DY / Chaitanya Vedithi S / Afanasyev P / Melero R / Warshamanage R / ...Authors: Matusevicius M / Corey RA / Gragera M / Yamashita K / Sprenger T / Ungogo MA / Blaza JN / Castro-Hartmann P / Chirgadze DY / Chaitanya Vedithi S / Afanasyev P / Melero R / Warshamanage R / Gusach A / Carazo JM / Carrington M / Blundell TL / Murshudov G / Stansfeld P / Sansom M / de Koning HP / Tate CG / Weyand SN | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16863.map.gz emd_16863.map.gz | 8.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16863-v30.xml emd-16863-v30.xml emd-16863.xml emd-16863.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16863_fsc.xml emd_16863_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_16863.png emd_16863.png | 69.9 KB | ||
| Masks |  emd_16863_msk_1.map emd_16863_msk_1.map | 9.2 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16863.cif.gz emd-16863.cif.gz | 6.3 KB | ||
| Others |  emd_16863_half_map_1.map.gz emd_16863_half_map_1.map.gz emd_16863_half_map_2.map.gz emd_16863_half_map_2.map.gz | 8.3 MB 8.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16863 http://ftp.pdbj.org/pub/emdb/structures/EMD-16863 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16863 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16863 | HTTPS FTP |
-Related structure data
| Related structure data |  8ofyMC  8ofxC  8ofzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16863.map.gz / Format: CCP4 / Size: 9.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16863.map.gz / Format: CCP4 / Size: 9.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.042 Å | ||||||||||||||||||||||||||||||||||||
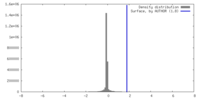
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16863_msk_1.map emd_16863_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
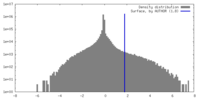
| Density Histograms |
-Half map: #2
| File | emd_16863_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
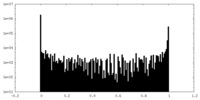
| Density Histograms |
-Half map: #1
| File | emd_16863_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
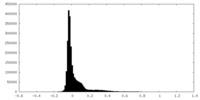
| Density Histograms |
- Sample components
Sample components
-Entire : Aquaporin 2 tetramer wildtype
| Entire | Name: Aquaporin 2 tetramer wildtype |
|---|---|
| Components |
|
-Supramolecule #1: Aquaporin 2 tetramer wildtype
| Supramolecule | Name: Aquaporin 2 tetramer wildtype / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 220 KDa |
-Macromolecule #1: Aquaglyceroporin 2
| Macromolecule | Name: Aquaglyceroporin 2 / type: protein_or_peptide / ID: 1 / Details: Pentamidine / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33.614645 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQSQPDNVAY PMELQAVNKD GTVEVRVQGN VDNSSNERWD ADVQKHEVAE AQEKPVGGIN FWAPRELRLN YRDYVAEFLG NFVLIYIAK GAVITSLLVP DFGLLGLTIG IGVAVTMALY VSLGISGGHL NSAVTVGNAV FGDFPWRKVP GYIAAQMLGT F LGAACAYG ...String: MQSQPDNVAY PMELQAVNKD GTVEVRVQGN VDNSSNERWD ADVQKHEVAE AQEKPVGGIN FWAPRELRLN YRDYVAEFLG NFVLIYIAK GAVITSLLVP DFGLLGLTIG IGVAVTMALY VSLGISGGHL NSAVTVGNAV FGDFPWRKVP GYIAAQMLGT F LGAACAYG VFADLLKAHG GGELIAFGEK GIAWVFAMYP AEGNGIFYPI FAELISTAVL LLCVCGIFDP NNSPAKGYET VA IGALVFV MVNNFGLASP LAMNPSLDFG PRVFGAILLG GEVFSHANYY FWVPLVVPFF GAILGLFLYK YFLPH UniProtKB: Aquaglyceroporin-2 |
-Macromolecule #2: 1,5-BIS(4-AMIDINOPHENOXY)PENTANE
| Macromolecule | Name: 1,5-BIS(4-AMIDINOPHENOXY)PENTANE / type: ligand / ID: 2 / Number of copies: 1 / Formula: PNT |
|---|---|
| Molecular weight | Theoretical: 340.419 Da |
| Chemical component information |  ChemComp-PNT: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
Details: 20 mM HEPES 100 mM NaCl | |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | Monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 56.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL |
|---|---|
| Output model |  PDB-8ofy: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)