+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the yeast SPT-Orm1-Dimer complex | |||||||||
 Map data Map data | Map-Dimer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Serine-Palmitoyl-Transferase / SPT / Orm-Protein / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of sphingolipid biosynthetic process / positive regulation of sphingolipid biosynthetic process / 3-keto-sphinganine metabolic process / intracellular sphingolipid homeostasis / serine palmitoyltransferase complex / serine C-palmitoyltransferase activity / serine C-palmitoyltransferase / ceramide metabolic process / sphingosine biosynthetic process / sphingolipid biosynthetic process ...negative regulation of sphingolipid biosynthetic process / positive regulation of sphingolipid biosynthetic process / 3-keto-sphinganine metabolic process / intracellular sphingolipid homeostasis / serine palmitoyltransferase complex / serine C-palmitoyltransferase activity / serine C-palmitoyltransferase / ceramide metabolic process / sphingosine biosynthetic process / sphingolipid biosynthetic process / ceramide biosynthetic process / response to unfolded protein / Neutrophil degranulation / enzyme activator activity / pyridoxal phosphate binding / endoplasmic reticulum membrane / endoplasmic reticulum / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Schaefer J / Koerner C / Parey K / Januliene D / Moeller A / Froehlich F | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure of the ceramide-bound SPOTS complex. Authors: Jan-Hannes Schäfer / Carolin Körner / Bianca M Esch / Sergej Limar / Kristian Parey / Stefan Walter / Dovile Januliene / Arne Moeller / Florian Fröhlich /  Abstract: Sphingolipids are structural membrane components that also function in cellular stress responses. The serine palmitoyltransferase (SPT) catalyzes the rate-limiting step in sphingolipid biogenesis. ...Sphingolipids are structural membrane components that also function in cellular stress responses. The serine palmitoyltransferase (SPT) catalyzes the rate-limiting step in sphingolipid biogenesis. Its activity is tightly regulated through multiple binding partners, including Tsc3, Orm proteins, ceramides, and the phosphatidylinositol-4-phosphate (PI4P) phosphatase Sac1. The structural organization and regulatory mechanisms of this complex are not yet understood. Here, we report the high-resolution cryo-EM structures of the yeast SPT in complex with Tsc3 and Orm1 (SPOT) as dimers and monomers and a monomeric complex further carrying Sac1 (SPOTS). In all complexes, the tight interaction of the downstream metabolite ceramide and Orm1 reveals the ceramide-dependent inhibition. Additionally, observation of ceramide and ergosterol binding suggests a co-regulation of sphingolipid biogenesis and sterol metabolism within the SPOTS complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16469.map.gz emd_16469.map.gz | 290.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16469-v30.xml emd-16469-v30.xml emd-16469.xml emd-16469.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16469_fsc.xml emd_16469_fsc.xml emd_16469_fsc_2.xml emd_16469_fsc_2.xml | 14.3 KB 14.3 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_16469.png emd_16469.png | 84.8 KB | ||
| Filedesc metadata |  emd-16469.cif.gz emd-16469.cif.gz | 7 KB | ||
| Others |  emd_16469_half_map_1.map.gz emd_16469_half_map_1.map.gz emd_16469_half_map_2.map.gz emd_16469_half_map_2.map.gz | 284.7 MB 284.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16469 http://ftp.pdbj.org/pub/emdb/structures/EMD-16469 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16469 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16469 | HTTPS FTP |
-Validation report
| Summary document |  emd_16469_validation.pdf.gz emd_16469_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16469_full_validation.pdf.gz emd_16469_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_16469_validation.xml.gz emd_16469_validation.xml.gz | 22.5 KB | Display | |
| Data in CIF |  emd_16469_validation.cif.gz emd_16469_validation.cif.gz | 28.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16469 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16469 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16469 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16469 | HTTPS FTP |
-Related structure data
| Related structure data |  8c82MC  8c80C  8c81C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16469.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16469.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map-Dimer | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.924 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half-A-Map-Dimer
| File | emd_16469_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-A-Map-Dimer | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-B-Map-Dimer
| File | emd_16469_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-B-Map-Dimer | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of Orm1 with Lcb1, Lcb2 and Tsc3
| Entire | Name: Complex of Orm1 with Lcb1, Lcb2 and Tsc3 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of Orm1 with Lcb1, Lcb2 and Tsc3
| Supramolecule | Name: Complex of Orm1 with Lcb1, Lcb2 and Tsc3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: C2-symmetric complex of Orm1 with Lcb1, Lcb2 and Tsc3 in each protomer |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 325 KDa |
-Macromolecule #1: Protein ORM1
| Macromolecule | Name: Protein ORM1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.221674 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTELDYQGTA EAASTSYSRN QTDLKPFPSA GSASSSIKTT EPVKDHRRRR AAAIISHVEP ETFEDENDQQ LLPNMNATWV DQRGAWIIH VVIIILLKLF YNLFPGVTTE WSWTLTNMTY VIGSYVMFHL IKGTPFDFNG GAYDNLTMWE QIDDETLYTP S RKFLISVP ...String: MTELDYQGTA EAASTSYSRN QTDLKPFPSA GSASSSIKTT EPVKDHRRRR AAAIISHVEP ETFEDENDQQ LLPNMNATWV DQRGAWIIH VVIIILLKLF YNLFPGVTTE WSWTLTNMTY VIGSYVMFHL IKGTPFDFNG GAYDNLTMWE QIDDETLYTP S RKFLISVP IALFLVSTHY AHYDLKLFSW NCFLTTFGAV VPKLPVTHRL RISIPGITGR AQIS UniProtKB: Protein ORM1 |
-Macromolecule #2: Serine palmitoyltransferase 1
| Macromolecule | Name: Serine palmitoyltransferase 1 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number: serine C-palmitoyltransferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 62.266793 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAHIPEVLPK SIPIPAFIVT TSSYLWYYFN LVLTQIPGGQ FIVSYIKKSH HDDPYRTTVE IGLILYGIIY YLSKPQQKKS LQAQKPNLS PQEIDALIED WEPEPLVDPS ATDEQSWRVA KTPVTMEMPI QNHITITRNN LQEKYTNVFN LASNNFLQLS A TEPVKEVV ...String: MAHIPEVLPK SIPIPAFIVT TSSYLWYYFN LVLTQIPGGQ FIVSYIKKSH HDDPYRTTVE IGLILYGIIY YLSKPQQKKS LQAQKPNLS PQEIDALIED WEPEPLVDPS ATDEQSWRVA KTPVTMEMPI QNHITITRNN LQEKYTNVFN LASNNFLQLS A TEPVKEVV KTTIKNYGVG ACGPAGFYGN QDVHYTLEYD LAQFFGTQGS VLYGQDFCAA PSVLPAFTKR GDVIVADDQV SL PVQNALQ LSRSTVYYFN HNDMNSLECL LNELTEQEKL EKLPAIPRKF IVTEGIFHNS GDLAPLPELT KLKNKYKFRL FVD ETFSIG VLGATGRGLS EHFNMDRATA IDITVGSMAT ALGSTGGFVL GDSVMCLHQR IGSNAYCFSA CLPAYTVTSV SKVL KLMDS NNDAVQTLQK LSKSLHDSFA SDDSLRSYVI VTSSPVSAVL HLQLTPAYRS RKFGYTCEQL FETMSALQKK SQTNK FIEP YEEEEKFLQS IVDHALINYN VLITRNTIVL KQETLPIVPS LKICCNAAMS PEELKNACES VKQSILACCQ ESNK UniProtKB: Serine palmitoyltransferase 1 |
-Macromolecule #3: Serine palmitoyltransferase 2
| Macromolecule | Name: Serine palmitoyltransferase 2 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO / EC number: serine C-palmitoyltransferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 63.189707 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSTPANYTRV PLCEPEELPD DIQKENEYGT LDSPGHLYQV KSRHGKPLPE PVVDTPPYYI SLLTYLNYLI LIILGHVHDF LGMTFQKNK HLDLLEHDGL APWFSNFESF YVRRIKMRID DCFSRPTTGV PGRFIRCIDR ISHNINEYFT YSGAVYPCMN L SSYNYLGF ...String: MSTPANYTRV PLCEPEELPD DIQKENEYGT LDSPGHLYQV KSRHGKPLPE PVVDTPPYYI SLLTYLNYLI LIILGHVHDF LGMTFQKNK HLDLLEHDGL APWFSNFESF YVRRIKMRID DCFSRPTTGV PGRFIRCIDR ISHNINEYFT YSGAVYPCMN L SSYNYLGF AQSKGQCTDA ALESVDKYSI QSGGPRAQIG TTDLHIKAEK LVARFIGKED ALVFSMGYGT NANLFNAFLD KK CLVISDE LNHTSIRTGV RLSGAAVRTF KHGDMVGLEK LIREQIVLGQ PKTNRPWKKI LICAEGLFSM EGTLCNLPKL VEL KKKYKC YLFIDEAHSI GAMGPTGRGV CEIFGVDPKD VDILMGTFTK SFGAAGGYIA ADQWIIDRLR LDLTTVSYSE SMPA PVLAQ TISSLQTISG EICPGQGTER LQRIAFNSRY LRLALQRLGF IVYGVADSPV IPLLLYCPSK MPAFSRMMLQ RRIAV VVVA YPATPLIESR VRFCMSASLT KEDIDYLLRH VSEVGDKLNL KSNSGKSSYD GKRQRWDIEE VIRRTPEDCK DDKYFV N UniProtKB: Serine palmitoyltransferase 2 |
-Macromolecule #4: Serine palmitoyltransferase-regulating protein TSC3
| Macromolecule | Name: Serine palmitoyltransferase-regulating protein TSC3 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 9.590233 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTQHKSSMVY IPTTKEAKRR NGKSEGILNT IEEVVEKLYW TYYIHLPFYL MASFDSFFLH VFFLTIFSLS FFGILKYCFL UniProtKB: Serine palmitoyltransferase-regulating protein TSC3 |
-Macromolecule #5: N-[(2S,3S,4R)-1,3,4-trihydroxyoctadecan-2-yl]hexacosanamide
| Macromolecule | Name: N-[(2S,3S,4R)-1,3,4-trihydroxyoctadecan-2-yl]hexacosanamide type: ligand / ID: 5 / Number of copies: 2 / Formula: Z8A |
|---|---|
| Molecular weight | Theoretical: 696.182 Da |
| Chemical component information | 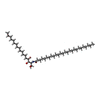 ChemComp-Z8A: |
-Macromolecule #6: PYRIDOXAL-5'-PHOSPHATE
| Macromolecule | Name: PYRIDOXAL-5'-PHOSPHATE / type: ligand / ID: 6 / Number of copies: 2 / Formula: PLP |
|---|---|
| Molecular weight | Theoretical: 247.142 Da |
| Chemical component information |  ChemComp-PLP: |
-Macromolecule #7: 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl...
| Macromolecule | Name: 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside type: ligand / ID: 7 / Number of copies: 2 / Formula: Q7G |
|---|---|
| Molecular weight | Theoretical: 1.165315 KDa |
| Chemical component information |  ChemComp-Q7G: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10.0 mg/mL |
|---|---|
| Buffer | pH: 6.8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number real images: 13604 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 105 |
|---|---|
| Output model |  PDB-8c82: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)