[English] 日本語
 Yorodumi
Yorodumi- EMDB-16426: CryoEM structure of the Hendra henipavirus nucleocapsid sauronoid... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of the Hendra henipavirus nucleocapsid sauronoid assembly multimer | |||||||||
 Map data Map data | Hendra henipavirus nucleoprotein sauronoid EM density postprocessed map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nucleoprotein / RNA-binding protein / Sauronoid / VIRAL PROTEIN | |||||||||
| Function / homology | Paramyxovirus nucleocapsid protein / Paramyxovirus nucleocapsid protein / helical viral capsid / viral nucleocapsid / host cell cytoplasm / ribonucleoprotein complex / structural molecule activity / RNA binding / Nucleocapsid Function and homology information Function and homology information | |||||||||
| Biological species |  Hendra henipavirus / Hendra henipavirus /  | |||||||||
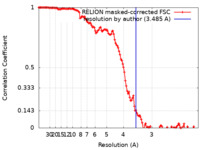
| Method | single particle reconstruction / cryo EM / Resolution: 3.485 Å | |||||||||
 Authors Authors | Passchier TC / Maskell DP / Edwards TA / Barr JN | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2024 Journal: Sci Rep / Year: 2024Title: The cryoEM structure of the Hendra henipavirus nucleoprotein reveals insights into paramyxoviral nucleocapsid architectures. Authors: Tim C Passchier / Joshua B R White / Daniel P Maskell / Matthew J Byrne / Neil A Ranson / Thomas A Edwards / John N Barr /   Abstract: We report the first cryoEM structure of the Hendra henipavirus nucleoprotein in complex with RNA, at 3.5 Å resolution, derived from single particle analysis of a double homotetradecameric RNA-bound ...We report the first cryoEM structure of the Hendra henipavirus nucleoprotein in complex with RNA, at 3.5 Å resolution, derived from single particle analysis of a double homotetradecameric RNA-bound N protein ring assembly exhibiting D14 symmetry. The structure of the HeV N protein adopts the common bi-lobed paramyxoviral N protein fold; the N-terminal and C-terminal globular domains are bisected by an RNA binding cleft containing six RNA nucleotides and are flanked by the N-terminal and C-terminal arms, respectively. In common with other paramyxoviral nucleocapsids, the lateral interface between adjacent N and N protomers involves electrostatic and hydrophobic interactions mediated primarily through the N-terminal arm and globular domains with minor contribution from the C-terminal arm. However, the HeV N multimeric assembly uniquely identifies an additional protomer-protomer contact between the N N-terminus and N C-terminal arm linker. The model presented here broadens the understanding of RNA-bound paramyxoviral nucleocapsid architectures and provides a platform for further insight into the molecular biology of HeV, as well as the development of antiviral interventions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16426.map.gz emd_16426.map.gz | 16.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16426-v30.xml emd-16426-v30.xml emd-16426.xml emd-16426.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16426_fsc.xml emd_16426_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_16426.png emd_16426.png | 117.6 KB | ||
| Masks |  emd_16426_msk_1.map emd_16426_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16426.cif.gz emd-16426.cif.gz | 6.2 KB | ||
| Others |  emd_16426_half_map_1.map.gz emd_16426_half_map_1.map.gz emd_16426_half_map_2.map.gz emd_16426_half_map_2.map.gz | 138.3 MB 138.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16426 http://ftp.pdbj.org/pub/emdb/structures/EMD-16426 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16426 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16426 | HTTPS FTP |
-Related structure data
| Related structure data |  8c4hMC  8cbwMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16426.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16426.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Hendra henipavirus nucleoprotein sauronoid EM density postprocessed map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.065 Å | ||||||||||||||||||||||||||||||||||||
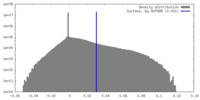
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16426_msk_1.map emd_16426_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
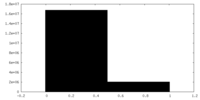
| Density Histograms |
-Half map: Hendra henipavirus nucleoprotein sauronoid EM density half map 2
| File | emd_16426_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Hendra henipavirus nucleoprotein sauronoid EM density half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
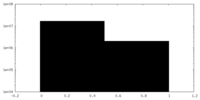
| Density Histograms |
-Half map: Hendra henipavirus nucleoprotein sauronoid EM density half map 1
| File | emd_16426_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Hendra henipavirus nucleoprotein sauronoid EM density half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
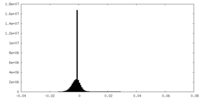
| Density Histograms |
- Sample components
Sample components
-Entire : Sauronoid assembly of Hendra henipavirus nucleoprotein
| Entire | Name: Sauronoid assembly of Hendra henipavirus nucleoprotein |
|---|---|
| Components |
|
-Supramolecule #1: Sauronoid assembly of Hendra henipavirus nucleoprotein
| Supramolecule | Name: Sauronoid assembly of Hendra henipavirus nucleoprotein type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Recombinantly expressed Hendra henipavirus nucleoprotein forming sauronoid complexes. |
|---|---|
| Source (natural) | Organism:  Hendra henipavirus Hendra henipavirus |
| Molecular weight | Theoretical: 1.691779 MDa |
-Macromolecule #1: Nucleocapsid
| Macromolecule | Name: Nucleocapsid / type: protein_or_peptide / ID: 1 / Number of copies: 28 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Hendra henipavirus Hendra henipavirus |
| Molecular weight | Theoretical: 58.54132 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSDIFDEAAS FRSYQSKLGR DGRASAATAT LTTKIRIFVP ATNSPELRWE LTLFALDVIR SPSAAESMKI GAAFTLISMY SERPGALIR SLLNDPDIEA VIIDVGSMLN GIPVMERRGD KAQEEMEGLM RILKTARESS KGKTPFVDSR AYGLRITDMS T LVSAVITI ...String: MSDIFDEAAS FRSYQSKLGR DGRASAATAT LTTKIRIFVP ATNSPELRWE LTLFALDVIR SPSAAESMKI GAAFTLISMY SERPGALIR SLLNDPDIEA VIIDVGSMLN GIPVMERRGD KAQEEMEGLM RILKTARESS KGKTPFVDSR AYGLRITDMS T LVSAVITI EAQIWILIAK AVTAPDTAEE SETRRWAKYV QQKRVNPFFA LTQQWLTEMR NLLSQSLSVR KFMVEILMEV KK GGSAKGR AVEIISDIGN YVEETGMAGF FATIRFGLET RYPALALNEF QSDLNTIKGL MLLYREIGPR APYMVLLEES IQT KFAPGG YPLLWSFAMG VATTIDRSMG ALNINRGYLE PMYFRLGQKS ARHHAGGIDQ NMANKLGLNS DQVAELAAAV QETS VGRQD NNMQAREAKF AAGGVLVGGG EQDIDEEEEP IEHSGRQSVT FKREMSMSSL ADSVPSSSVS TSGGTRLTNS LLNLR SRLA AKAIKESTAQ SSSERNPPNN RPQADSGRKD DQEPKPAQND LDFVRADV UniProtKB: Nucleocapsid |
-Macromolecule #2: RNA (84-MER)
| Macromolecule | Name: RNA (84-MER) / type: rna / ID: 2 / Number of copies: 2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.672984 KDa |
| Sequence | String: UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 3548 / Average exposure time: 1.5 sec. / Average electron dose: 59.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)