[English] 日本語
 Yorodumi
Yorodumi- EMDB-16325: Cryo-EM structure of SKP1-SKP2-CKS1-CDK2-CyclinA-p27KIP1 Complex -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SKP1-SKP2-CKS1-CDK2-CyclinA-p27KIP1 Complex | |||||||||
 Map data Map data | Full map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cell cycle / cyclin-dependent kinase / signalling / ubiquitination | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of protein polyubiquitination / negative regulation of growth / F-box domain binding / cellular response to cell-matrix adhesion / : / cyclin A2-CDK1 complex / Aberrant regulation of mitotic exit in cancer due to RB1 defects / PcG protein complex / cell cycle G1/S phase transition / cellular response to luteinizing hormone stimulus ...positive regulation of protein polyubiquitination / negative regulation of growth / F-box domain binding / cellular response to cell-matrix adhesion / : / cyclin A2-CDK1 complex / Aberrant regulation of mitotic exit in cancer due to RB1 defects / PcG protein complex / cell cycle G1/S phase transition / cellular response to luteinizing hormone stimulus / positive regulation of ubiquitin protein ligase activity / cyclin-dependent protein serine/threonine kinase inhibitor activity / Transcription of E2F targets under negative control by p107 (RBL1) and p130 (RBL2) in complex with HDAC1 / cellular response to leptin stimulus / programmed cell death / Cul7-RING ubiquitin ligase complex / maintenance of protein location in nucleus / Loss of Function of FBXW7 in Cancer and NOTCH1 Signaling / male pronucleus / cyclin-dependent protein serine/threonine kinase activator activity / female pronucleus / cellular response to cocaine / response to glucagon / cyclin-dependent protein serine/threonine kinase regulator activity / positive regulation of DNA biosynthetic process / SCF ubiquitin ligase complex / SCF-dependent proteasomal ubiquitin-dependent protein catabolic process / positive regulation of intracellular estrogen receptor signaling pathway / regulation of mitotic cell cycle phase transition / cellular response to insulin-like growth factor stimulus / cyclin A1-CDK2 complex / cyclin E2-CDK2 complex / cyclin E1-CDK2 complex / cyclin A2-CDK2 complex / positive regulation of DNA-templated DNA replication initiation / G2 Phase / Y chromosome / cyclin-dependent protein kinase activity / Phosphorylation of proteins involved in G1/S transition by active Cyclin E:Cdk2 complexes / positive regulation of heterochromatin formation / p53-Dependent G1 DNA Damage Response / X chromosome / Prolactin receptor signaling / PTK6 Regulates Cell Cycle / ubiquitin ligase complex scaffold activity / regulation of anaphase-promoting complex-dependent catabolic process / Defective binding of RB1 mutants to E2F1,(E2F2, E2F3) / centriole replication / Regulation of APC/C activators between G1/S and early anaphase / telomere maintenance in response to DNA damage / regulation of DNA replication / microtubule organizing center / centrosome duplication / G0 and Early G1 / cullin family protein binding / cochlea development / animal organ regeneration / Telomere Extension By Telomerase / Activation of the pre-replicative complex / protein K63-linked ubiquitination / protein monoubiquitination / cyclin-dependent kinase / cyclin-dependent protein serine/threonine kinase activity / TP53 Regulates Transcription of Genes Involved in G1 Cell Cycle Arrest / Activation of ATR in response to replication stress / Regulation of MITF-M-dependent genes involved in cell cycle and proliferation / Cyclin E associated events during G1/S transition / Cajal body / ubiquitin-like ligase-substrate adaptor activity / protein K48-linked ubiquitination / Cyclin A:Cdk2-associated events at S phase entry / positive regulation of double-strand break repair via homologous recombination / Cyclin A/B1/B2 associated events during G2/M transition / cyclin-dependent protein kinase holoenzyme complex / Nuclear events stimulated by ALK signaling in cancer / regulation of G2/M transition of mitotic cell cycle / condensed chromosome / cellular response to platelet-derived growth factor stimulus / mitotic G1 DNA damage checkpoint signaling / positive regulation of smooth muscle cell proliferation / cellular response to nitric oxide / post-translational protein modification / regulation of mitotic cell cycle / molecular function activator activity / cyclin binding / positive regulation of DNA replication / Regulation of BACH1 activity / ubiquitin binding / MAP3K8 (TPL2)-dependent MAPK1/3 activation / male germ cell nucleus / meiotic cell cycle / SCF-beta-TrCP mediated degradation of Emi1 / NIK-->noncanonical NF-kB signaling / Vpu mediated degradation of CD4 / cellular response to estradiol stimulus / Cdc20:Phospho-APC/C mediated degradation of Cyclin A / Dectin-1 mediated noncanonical NF-kB signaling / Activation of NF-kappaB in B cells / Degradation of GLI1 by the proteasome / G1/S transition of mitotic cell cycle Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
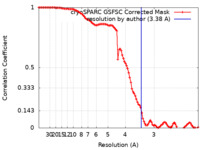
| Method | single particle reconstruction / cryo EM / Resolution: 3.38 Å | |||||||||
 Authors Authors | Rowland RJ / Salamina M / Endicott JA / Noble ME | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2023 Journal: Sci Rep / Year: 2023Title: Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1. Authors: Rhianna J Rowland / Richard Heath / Daniel Maskell / Rebecca F Thompson / Neil A Ranson / James N Blaza / Jane A Endicott / Martin E M Noble / Marco Salamina /  Abstract: p27KIP1 (cyclin-dependent kinase inhibitor 1B, p27) is a member of the CIP/KIP family of CDK (cyclin dependent kinase) regulators that inhibit cell cycle CDKs. p27 phosphorylation by CDK1/2, signals ...p27KIP1 (cyclin-dependent kinase inhibitor 1B, p27) is a member of the CIP/KIP family of CDK (cyclin dependent kinase) regulators that inhibit cell cycle CDKs. p27 phosphorylation by CDK1/2, signals its recruitment to the SCF (S-phase kinase associated protein 1 (SKP1)-cullin-SKP2) E3 ubiquitin ligase complex for proteasomal degradation. The nature of p27 binding to SKP2 and CKS1 was revealed by the SKP1-SKP2-CKS1-p27 phosphopeptide crystal structure. Subsequently, a model for the hexameric CDK2-cyclin A-CKS1-p27-SKP1-SKP2 complex was proposed by overlaying an independently determined CDK2-cyclin A-p27 structure. Here we describe the experimentally determined structure of the isolated CDK2-cyclin A-CKS1-p27-SKP1-SKP2 complex at 3.4 Å global resolution using cryogenic electron microscopy. This structure supports previous analysis in which p27 was found to be structurally dynamic, transitioning from disordered to nascent secondary structure on target binding. We employed 3D variability analysis to further explore the conformational space of the hexameric complex and uncovered a previously unidentified hinge motion centred on CKS1. This flexibility gives rise to open and closed conformations of the hexameric complex that we propose may contribute to p27 regulation by facilitating recognition with SCF. This 3D variability analysis further informed particle subtraction and local refinement approaches to enhance the local resolution of the complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16325.map.gz emd_16325.map.gz | 97.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16325-v30.xml emd-16325-v30.xml emd-16325.xml emd-16325.xml | 24.8 KB 24.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16325_fsc.xml emd_16325_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_16325.png emd_16325.png | 96.2 KB | ||
| Masks |  emd_16325_msk_1.map emd_16325_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16325.cif.gz emd-16325.cif.gz | 7.4 KB | ||
| Others |  emd_16325_half_map_1.map.gz emd_16325_half_map_1.map.gz emd_16325_half_map_2.map.gz emd_16325_half_map_2.map.gz | 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16325 http://ftp.pdbj.org/pub/emdb/structures/EMD-16325 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16325 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16325 | HTTPS FTP |
-Related structure data
| Related structure data |  8byaMC  8bylC  8bzoC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16325.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16325.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16325_msk_1.map emd_16325_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_16325_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B
| File | emd_16325_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Hexameric complex of SKP1-SKP2-CKS1 with CDK2-CyclinA-p27(kip1)
| Entire | Name: Hexameric complex of SKP1-SKP2-CKS1 with CDK2-CyclinA-p27(kip1) |
|---|---|
| Components |
|
-Supramolecule #1: Hexameric complex of SKP1-SKP2-CKS1 with CDK2-CyclinA-p27(kip1)
| Supramolecule | Name: Hexameric complex of SKP1-SKP2-CKS1 with CDK2-CyclinA-p27(kip1) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 175 KDa |
-Macromolecule #1: Cyclin-dependent kinase 2
| Macromolecule | Name: Cyclin-dependent kinase 2 / type: protein_or_peptide / ID: 1 / Details: Thr160 phosphorylated CDK2 / Number of copies: 1 / Enantiomer: LEVO / EC number: cyclin-dependent kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 33.994398 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MENFQKVEKI GEGTYGVVYK ARNKLTGEVV ALKKIRLDTE TEGVPSTAIR EISLLKELNH PNIVKLLDVI HTENKLYLVF EFLHQDLKK FMDASALTGI PLPLIKSYLF QLLQGLAFCH SHRVLHRDLK PQNLLINTEG AIKLADFGLA RAFGVPVRT (TPO)THEVVTLWY ...String: MENFQKVEKI GEGTYGVVYK ARNKLTGEVV ALKKIRLDTE TEGVPSTAIR EISLLKELNH PNIVKLLDVI HTENKLYLVF EFLHQDLKK FMDASALTGI PLPLIKSYLF QLLQGLAFCH SHRVLHRDLK PQNLLINTEG AIKLADFGLA RAFGVPVRT (TPO)THEVVTLWY RAPEILLGCK YYSTAVDIWS LGCIFAEMVT RRALFPGDSE IDQLFRIFRT LGTPDEVVWP GVTSMP DYK PSFPKWARQD FSKVVPPLDE DGRSLLSQML HYDPNKRISA KAALAHPFFQ DVTKPVPHLR L UniProtKB: Cyclin-dependent kinase 2 |
-Macromolecule #2: Cyclin-A2
| Macromolecule | Name: Cyclin-A2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 48.609574 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLGNSAPGPA TREAGSALLA LQQTALQEDQ ENINPEKAAP VQQPRTRAAL AVLKSGNPRG LAQQQRPKTR RVAPLKDLPV NDEHVTVPP WKANSKQPAF TIHVDEAEKE AQKKPAESQK IEREDALAFN SAISLPGPRK PLVPLDYPMD GSFESPHTMD M SIILEDEK ...String: MLGNSAPGPA TREAGSALLA LQQTALQEDQ ENINPEKAAP VQQPRTRAAL AVLKSGNPRG LAQQQRPKTR RVAPLKDLPV NDEHVTVPP WKANSKQPAF TIHVDEAEKE AQKKPAESQK IEREDALAFN SAISLPGPRK PLVPLDYPMD GSFESPHTMD M SIILEDEK PVSVNEVPDY HEDIHTYLRE MEVKCKPKVG YMKKQPDITN SMRAILVDWL VEVGEEYKLQ NETLHLAVNY ID RFLSSMS VLRGKLQLVG TAAMLLASKF EEIYPPEVAE FVYITDDTYT KKQVLRMEHL VLKVLTFDLA APTVNQFLTQ YFL HQQPAN CKVESLAMFL GELSLIDADP YLKYLPSVIA GAAFHLALYT VTGQSWPESL IRKTGYTLES LKPCLMDLHQ TYLK APQHA QQSIREKYKN SKYHGVSLLN PPETLNL UniProtKB: Cyclin-A2 |
-Macromolecule #3: Cyclin-dependent kinase inhibitor 1B
| Macromolecule | Name: Cyclin-dependent kinase inhibitor 1B / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 17.678531 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSNVRVSNGS PSLERMDARQ AEHPKPSACR NLFGPVDHEE LTRDLEKHCR DMEEASQRKW NFDFQNHKPL EGKYEWQEVE KGSLPEFYY RPPRPPKGAC KVPAQESQDG SGSRPAAPLI GAPANSEDTH LVDPKTDPSD SQTGLAEQCA GIRKRPATD UniProtKB: Cyclin-dependent kinase inhibitor 1B |
-Macromolecule #4: S-phase kinase-associated protein 1
| Macromolecule | Name: S-phase kinase-associated protein 1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 18.679965 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPSIKLQSSD GEIFEVDVEI AKQSVTIKTM LEDLGMDDEG DDDPVPLPNV NAAILKKVIQ WCTHHKDDPP PPEDDENKEK RTDDIPVWD QEFLKVDQGT LFELILAANY LDIKGLLDVT CKTVANMIKG KTPEEIRKTF NIKNDFTEEE EAQVRKENQW C EEK UniProtKB: S-phase kinase-associated protein 1 |
-Macromolecule #5: S-phase kinase-associated protein 2
| Macromolecule | Name: S-phase kinase-associated protein 2 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 47.817785 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHRKHLQEIP DLSSNVATSF TWGWDSSKTS ELLSGMGVSA LEKEEPDSEN IPQELLSNLG HPESPPRKRL KSKGSDKDFV IVRRPKLNR ENFPGVSWDS LPDELLLGIF SCLCLPELLK VSGVCKRWYR LASDESLWQT LDLTGKNLHP DVTGRLLSQG V IAFRCPRS ...String: MHRKHLQEIP DLSSNVATSF TWGWDSSKTS ELLSGMGVSA LEKEEPDSEN IPQELLSNLG HPESPPRKRL KSKGSDKDFV IVRRPKLNR ENFPGVSWDS LPDELLLGIF SCLCLPELLK VSGVCKRWYR LASDESLWQT LDLTGKNLHP DVTGRLLSQG V IAFRCPRS FMDQPLAEHF SPFRVQHMDL SNSVIEVSTL HGILSQCSKL QNLSLEGLRL SDPIVNTLAK NSNLVRLNLS GC SGFSEFA LQTLLSSCSR LDELNLSWCF DFTEKHVQVA VAHVSETITQ LNLSGYRKNL QKSDLSTLVR RCPNLVHLDL SDS VMLKND CFQEFFQLNY LQHLSLSRCY DIIPETLLEL GEIPTLKTLQ VFGIVPDGTL QLLKEALPHL QINCSHFTTI ARPT IGNKK NQEIWGIKCR LTLQKPSCL UniProtKB: S-phase kinase-associated protein 2 |
-Macromolecule #6: Cyclin-dependent kinases regulatory subunit 1
| Macromolecule | Name: Cyclin-dependent kinases regulatory subunit 1 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 9.679211 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSHKQIYYSD KYDDEEFEYR HVMLPKDIAK LVPKTHLMSE SEWRNLGVQQ SQGWVHYMIH EPEPHILLFR RPLPKKPKK UniProtKB: Cyclin-dependent kinases regulatory subunit 1 |
-Macromolecule #7: p27 KIP1 C-terminus
| Macromolecule | Name: p27 KIP1 C-terminus / type: protein_or_peptide / ID: 7 / Details: C-terminus of p27 KIP1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.126154 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AGSVEQ(TPO)PKK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 278.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average exposure time: 9.0 sec. / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Initial fitting was performed in chimera followed by real space refinement in Phenix |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 121 |
| Output model |  PDB-8bya: |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)